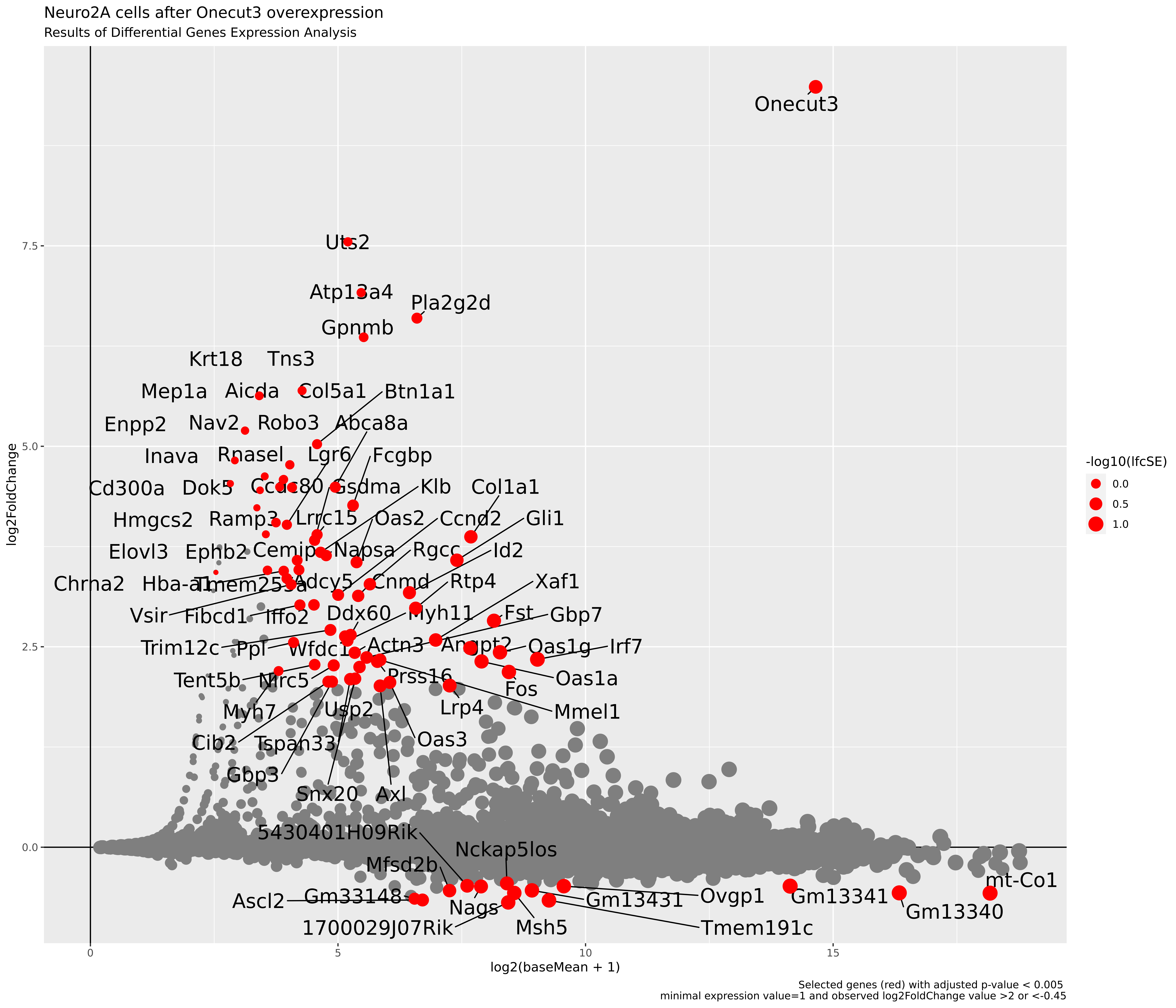
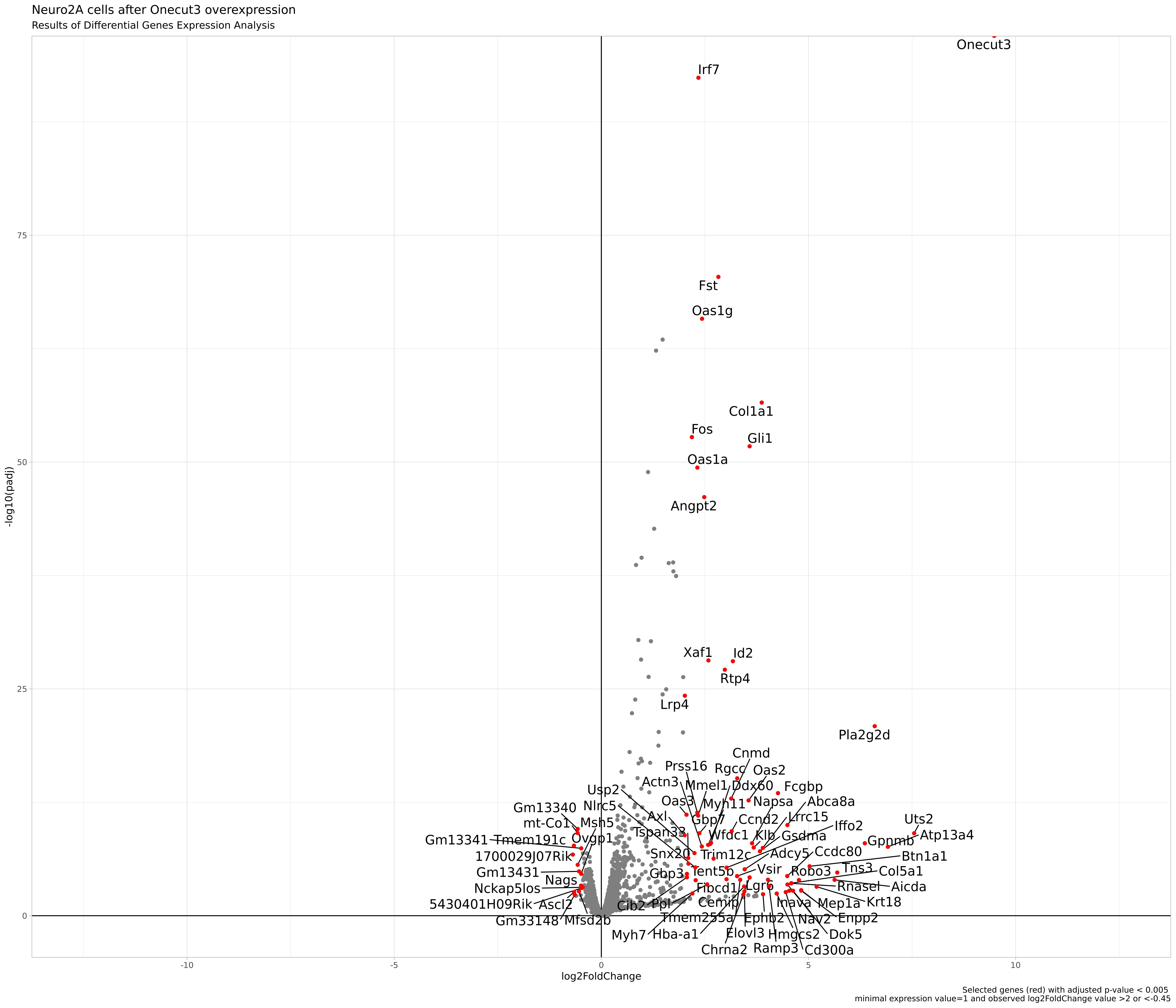
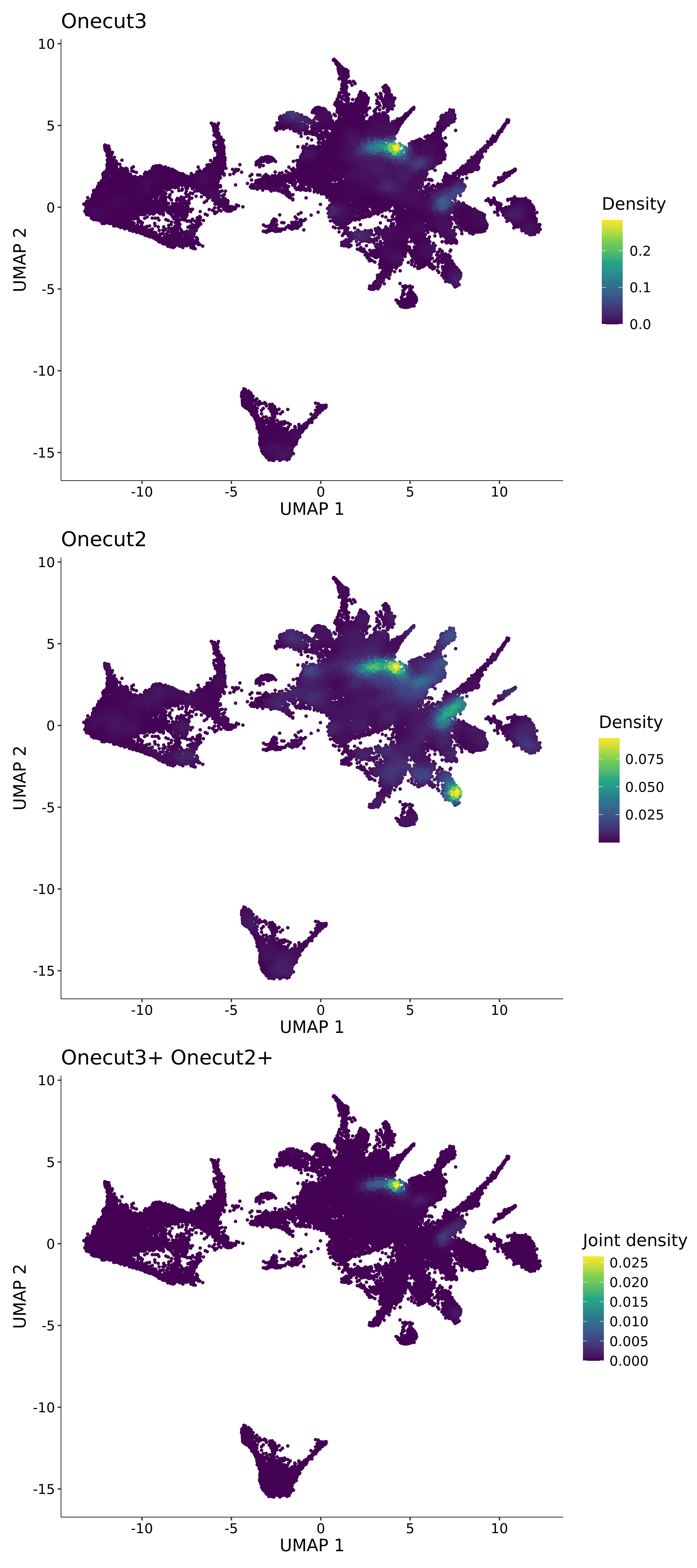
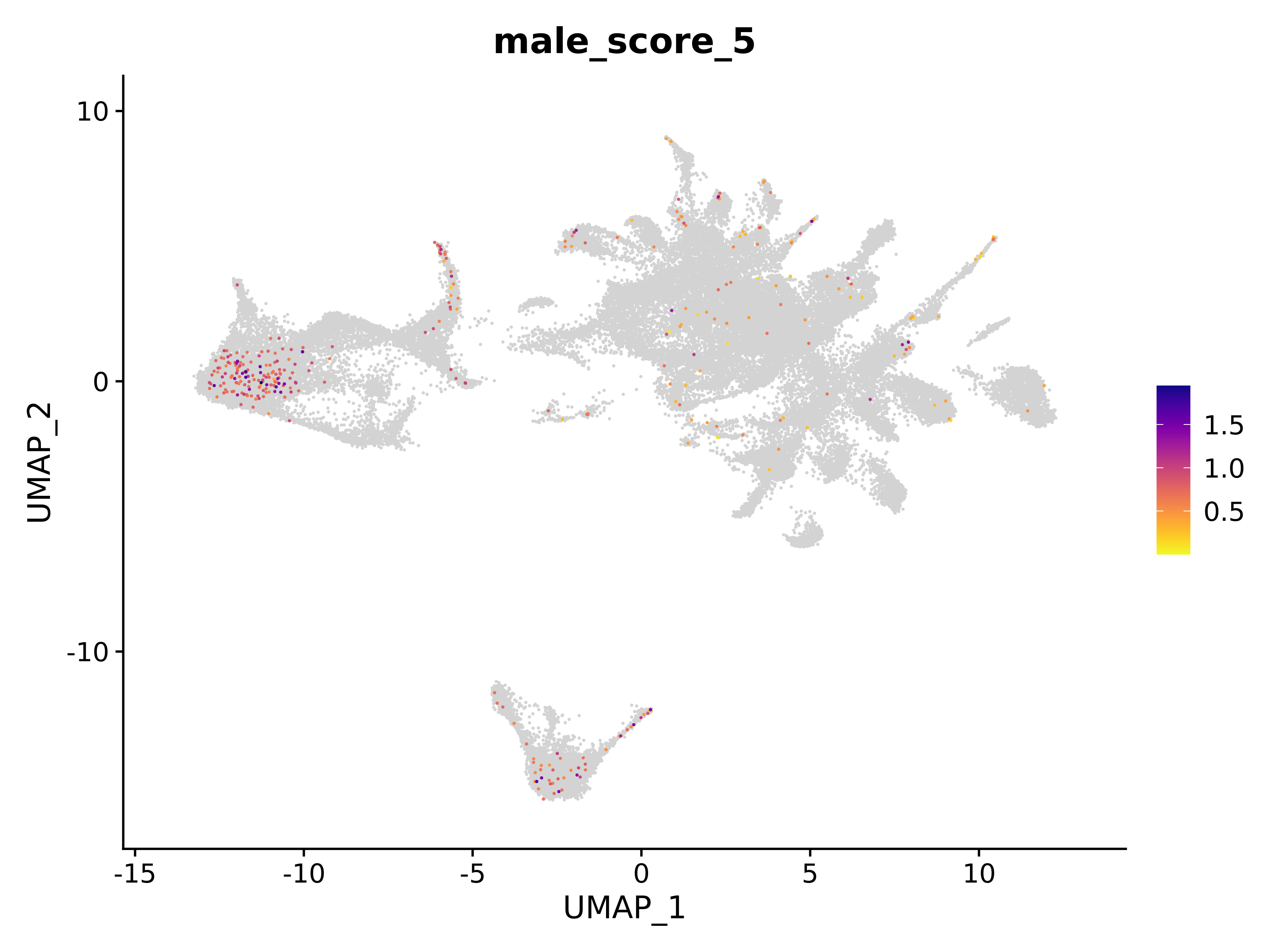
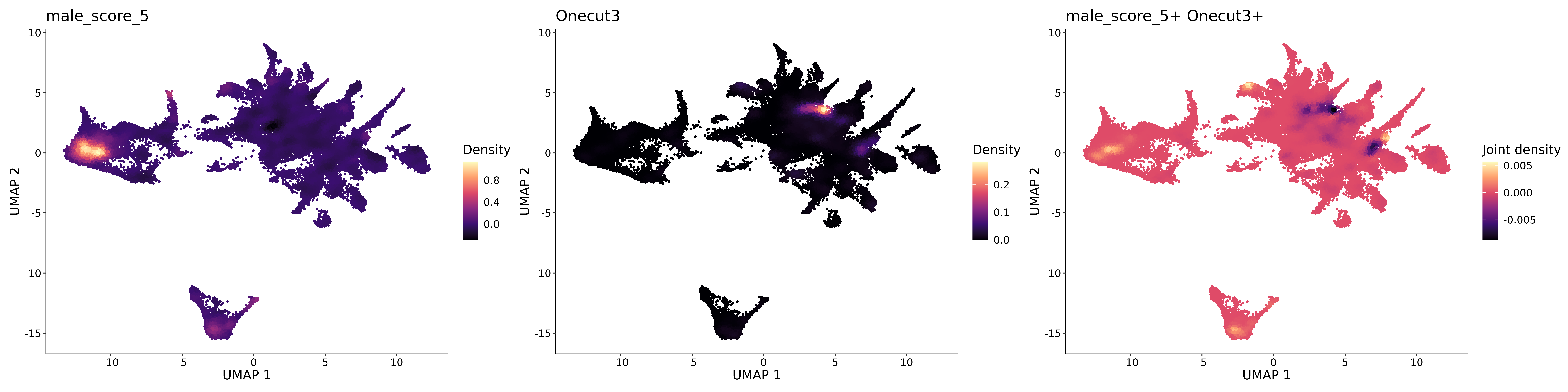
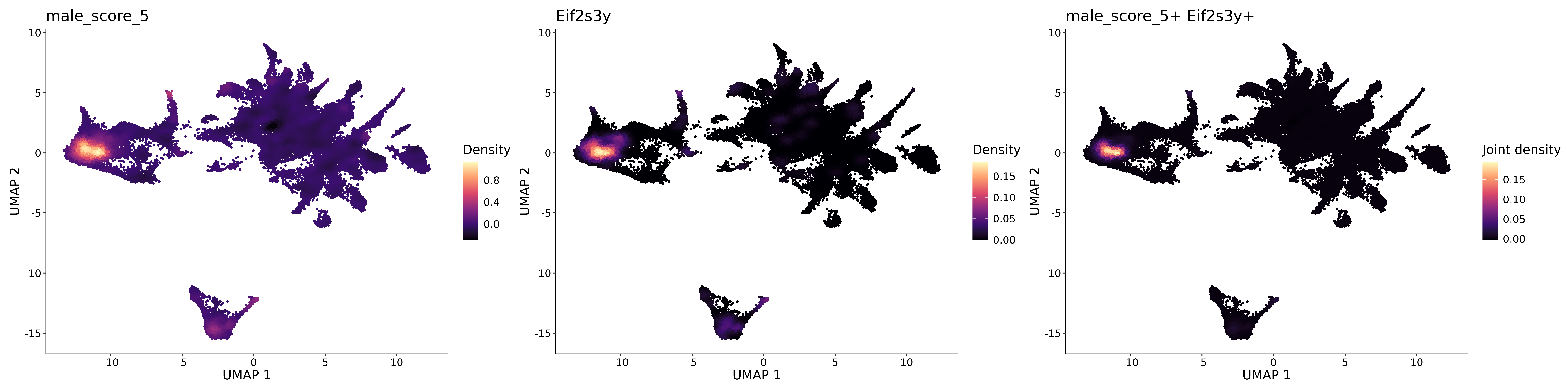
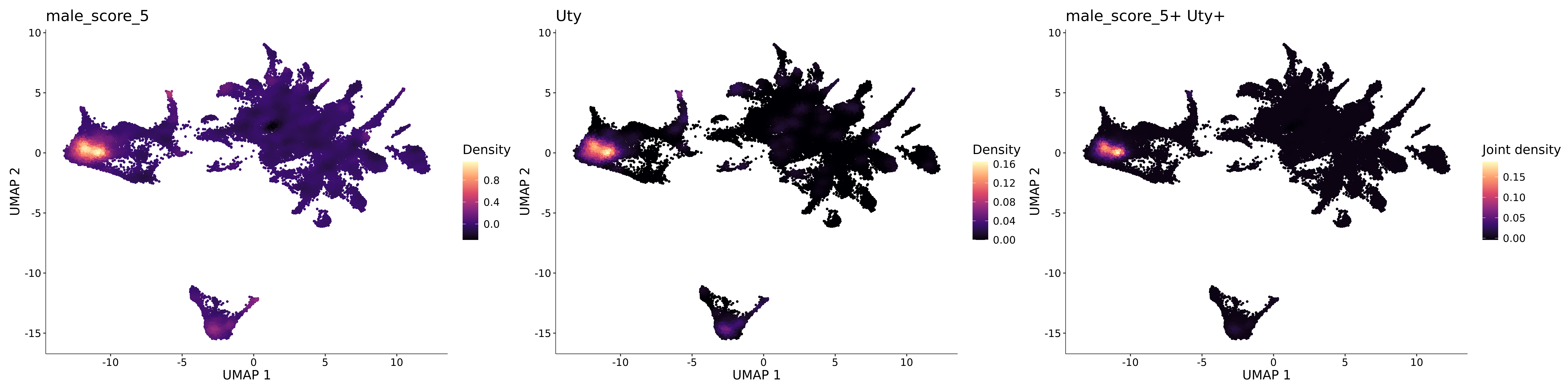
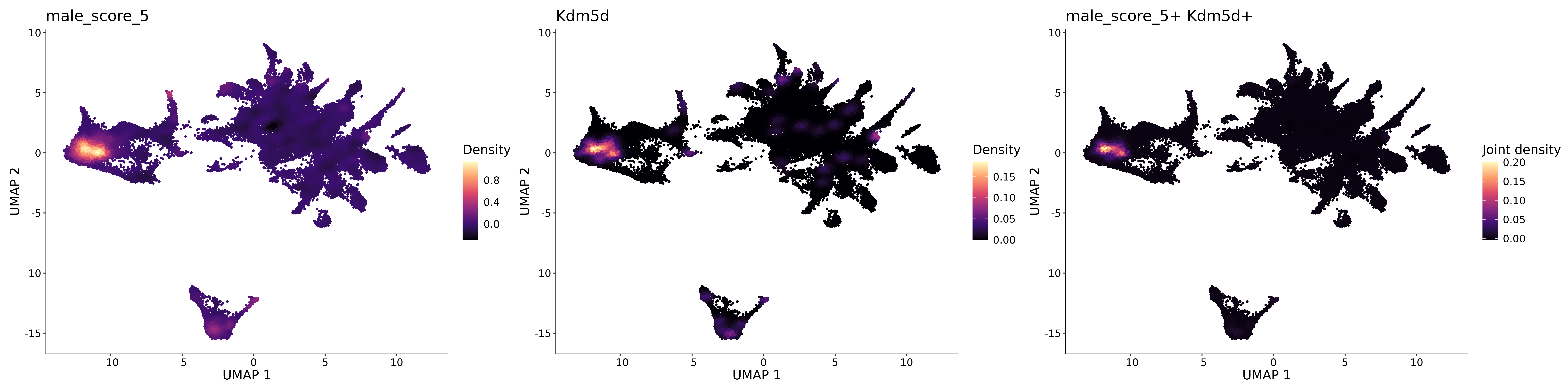
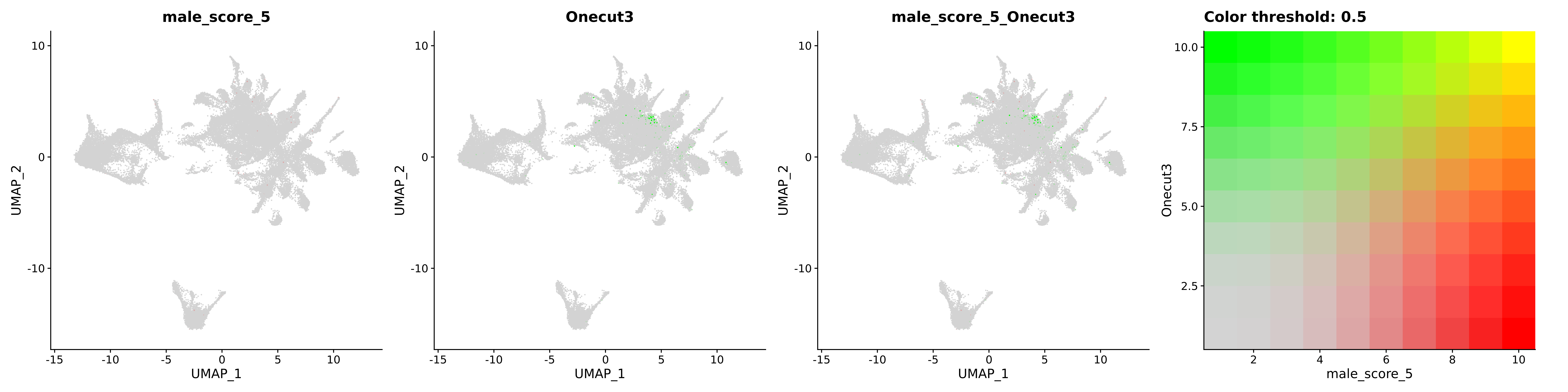
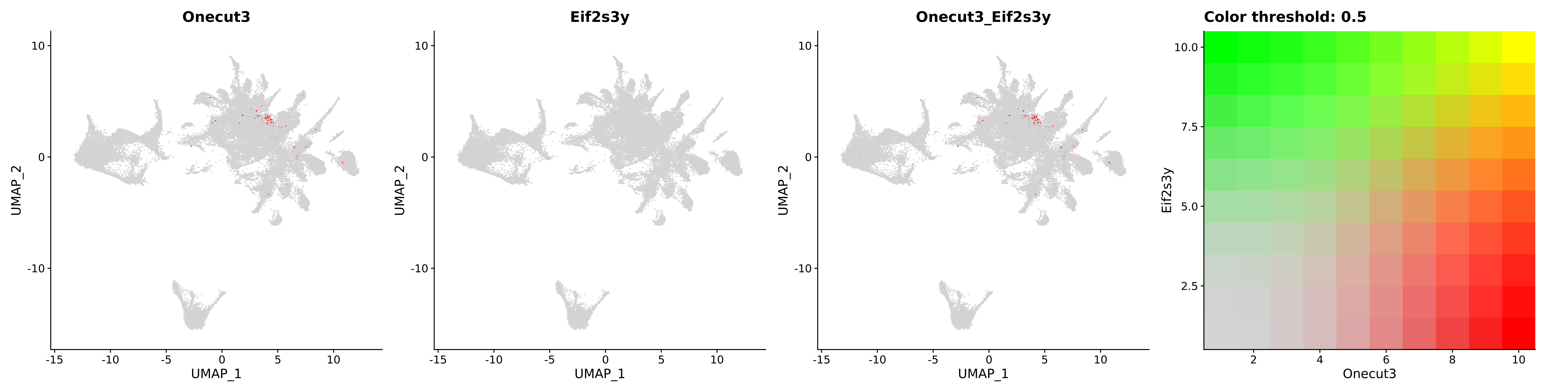
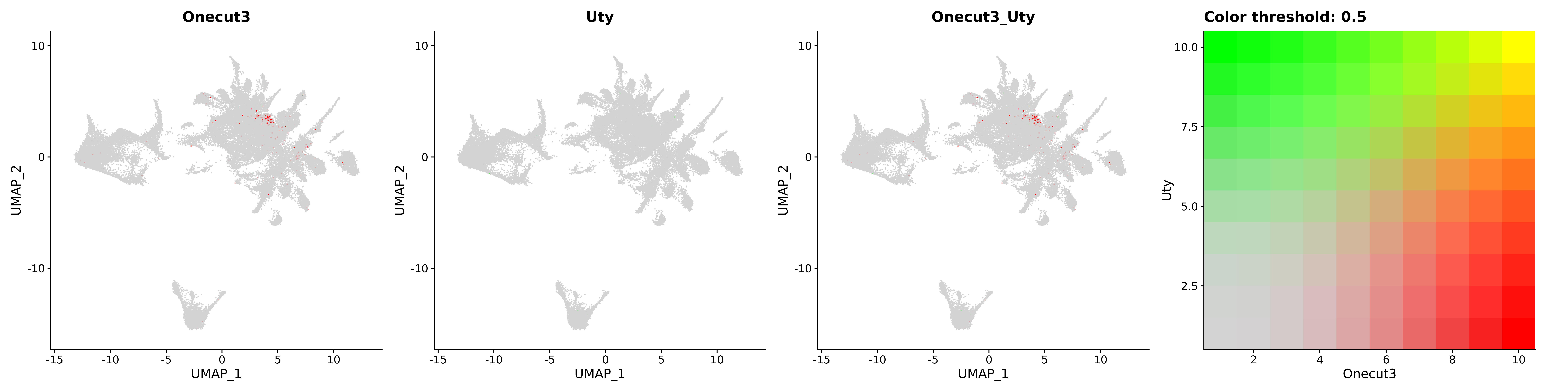
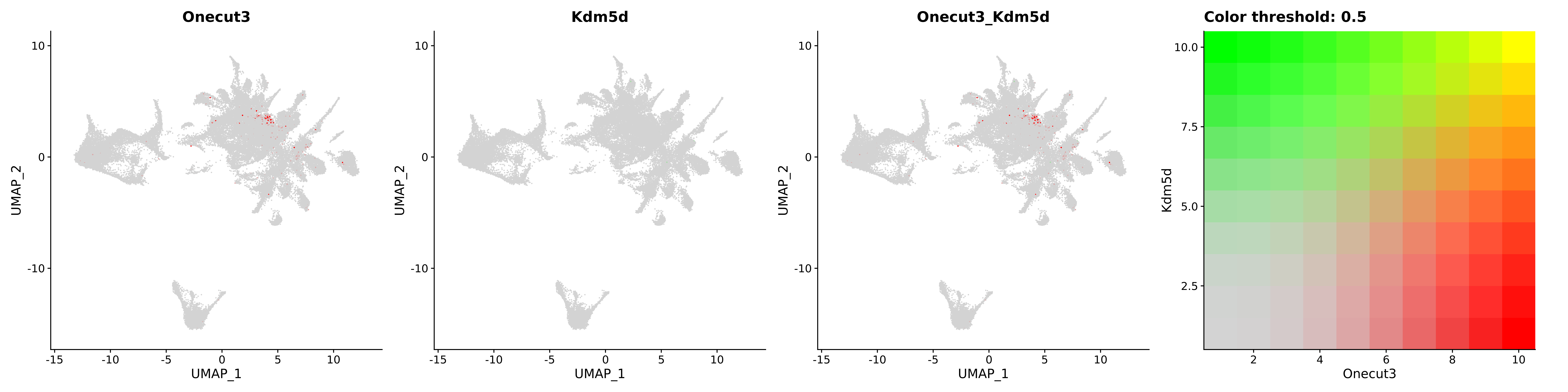
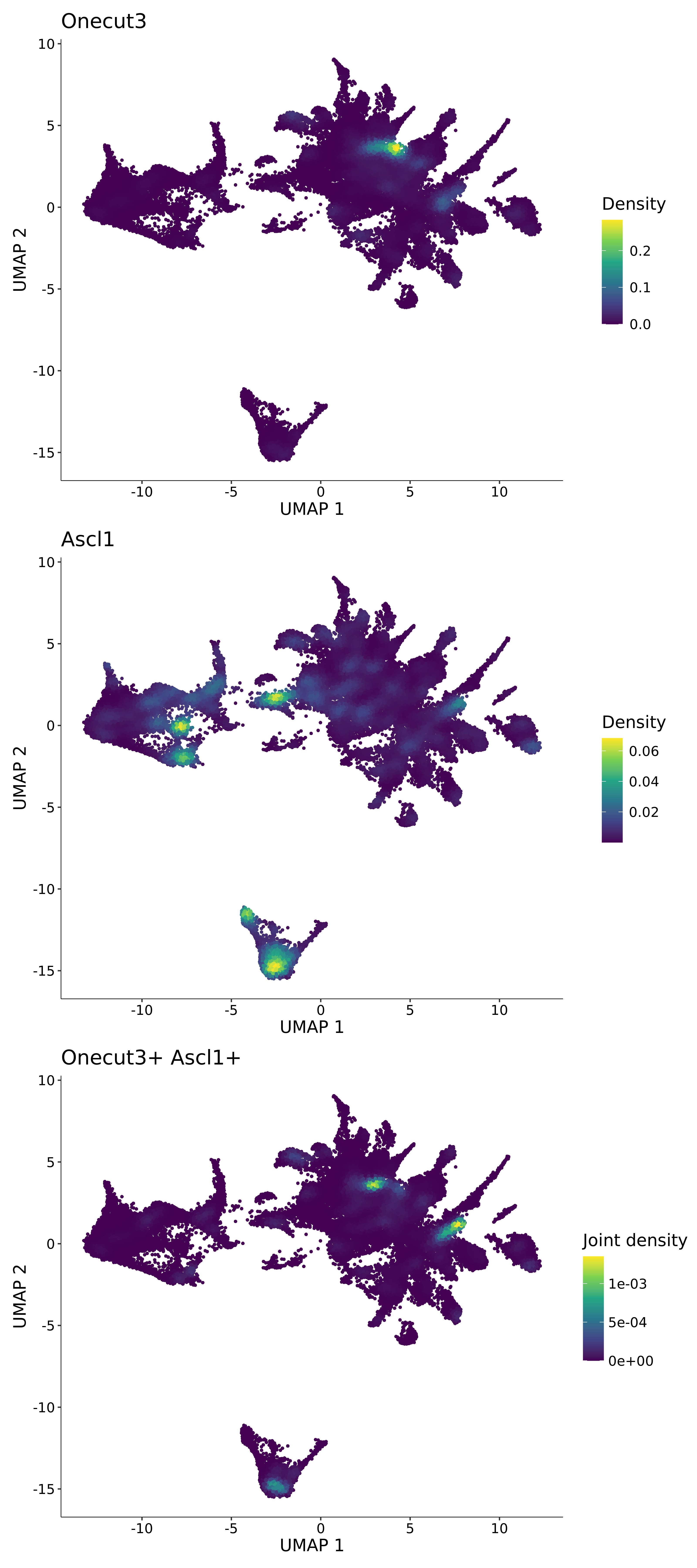
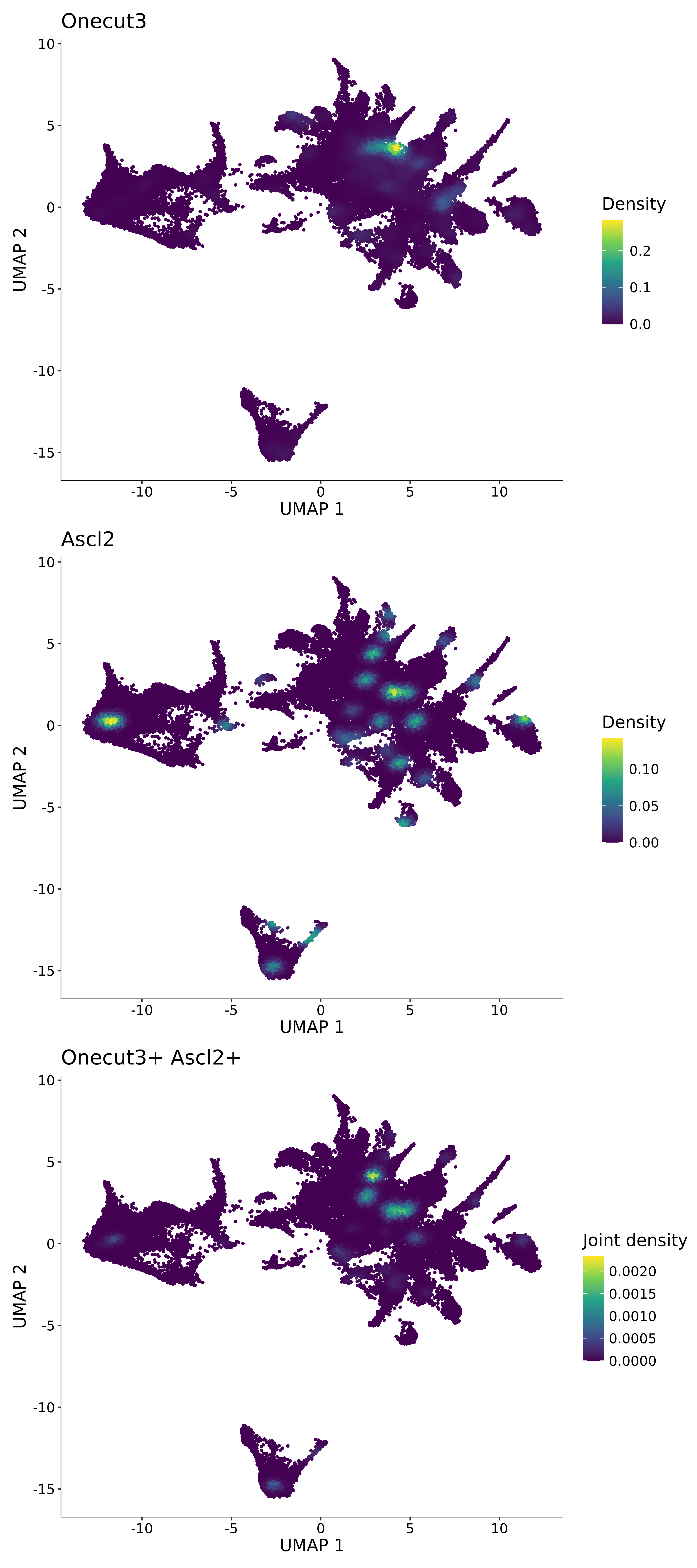
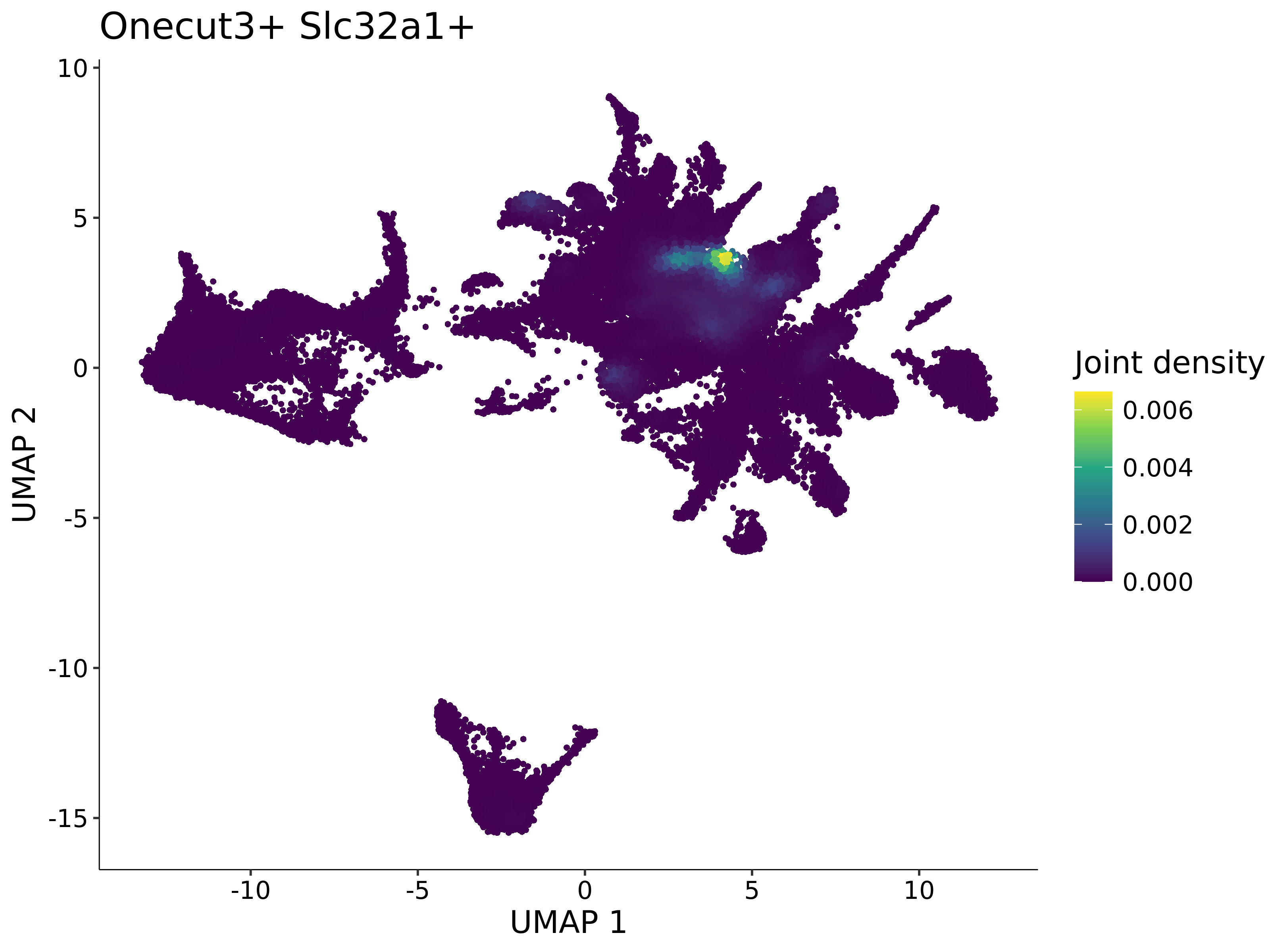
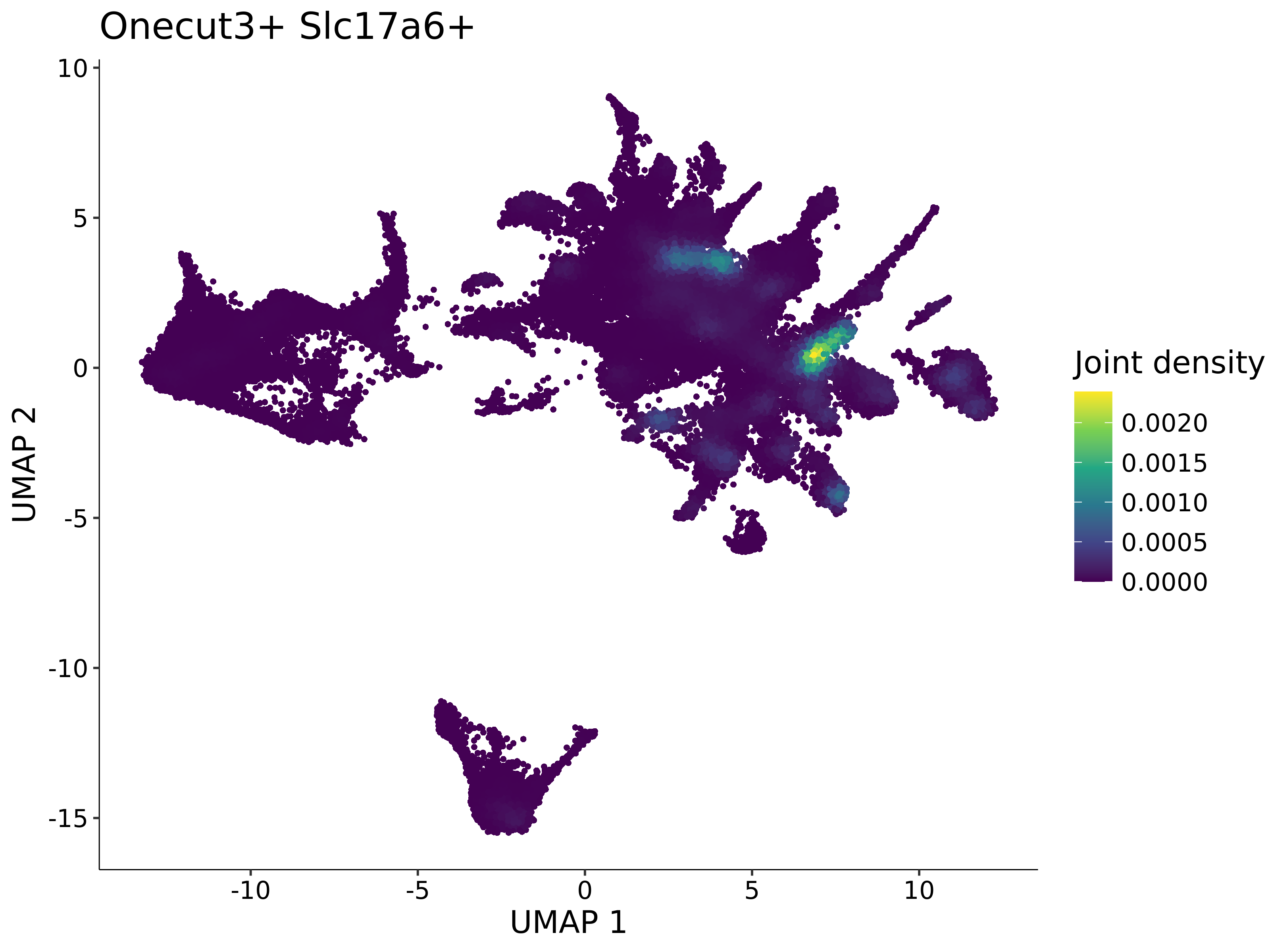
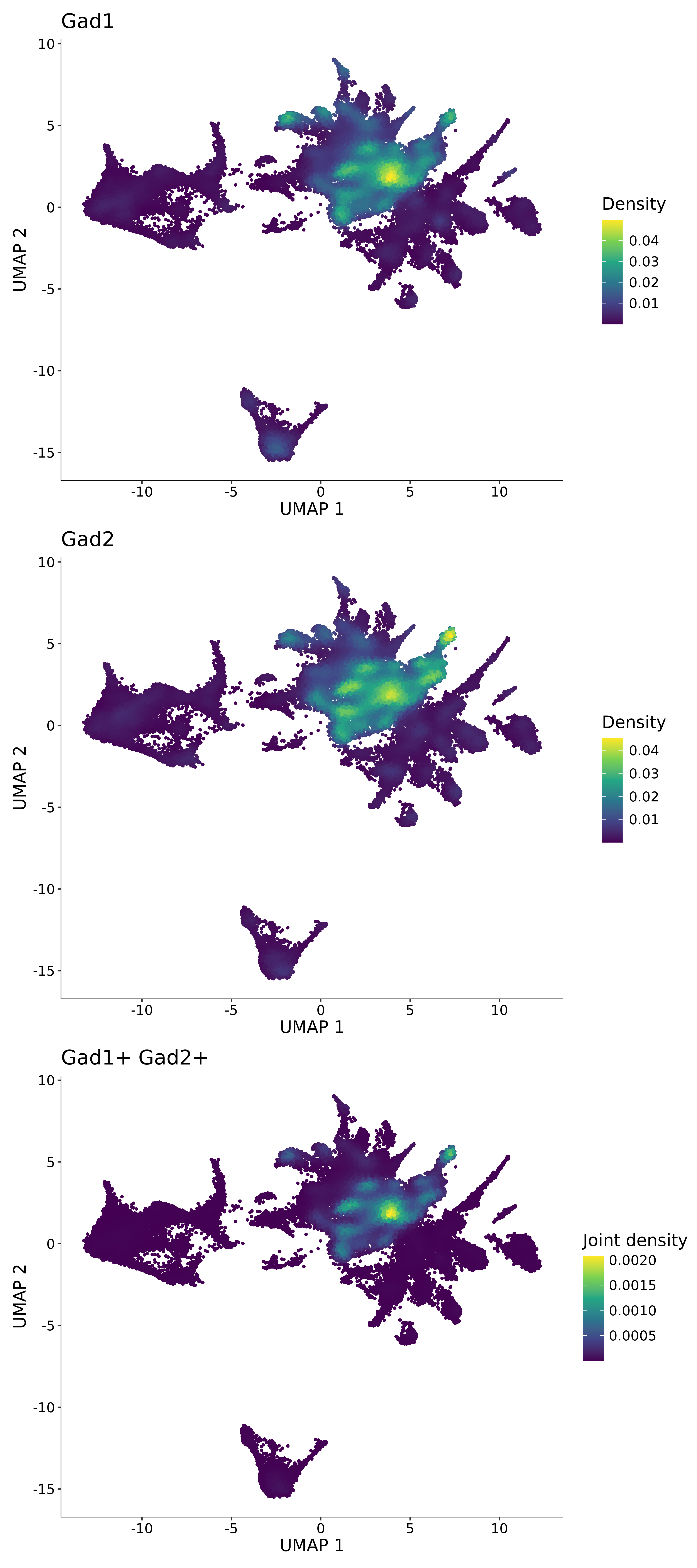
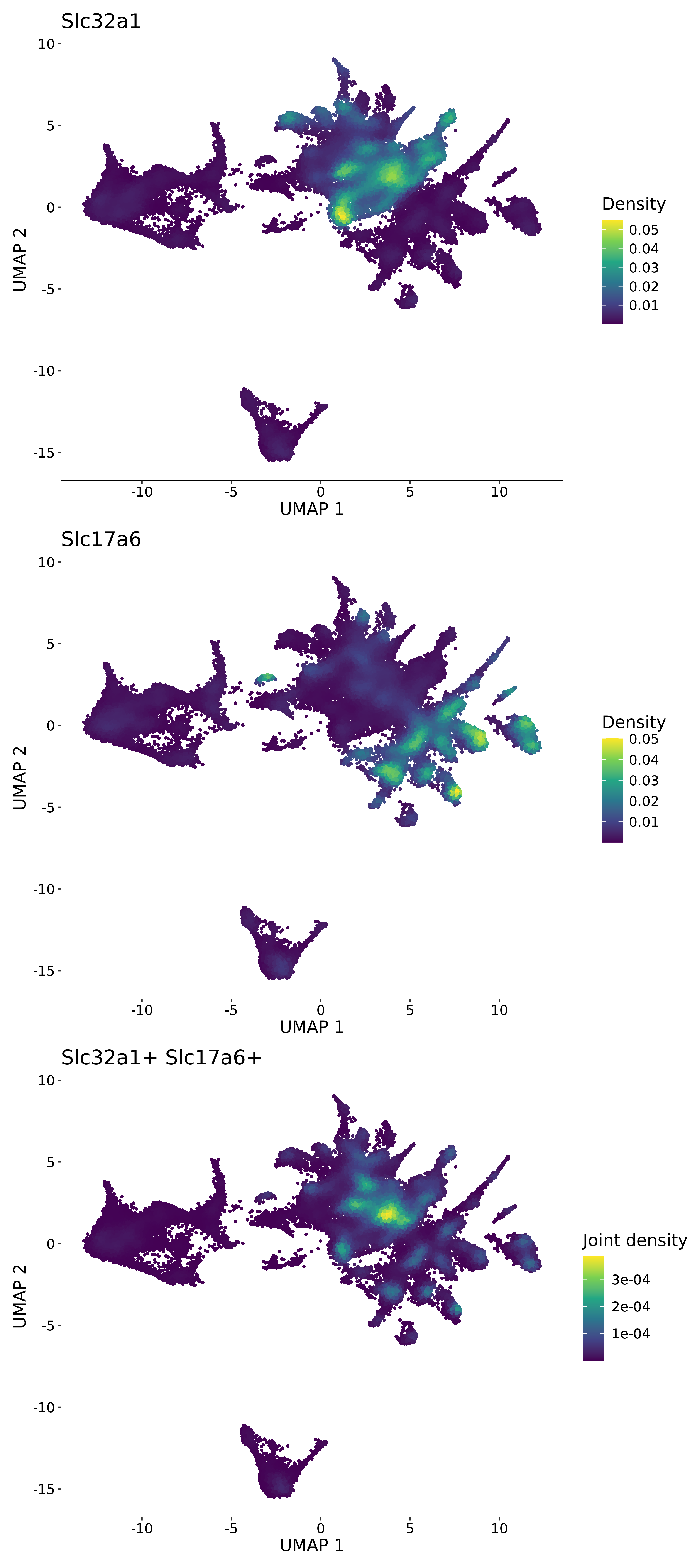
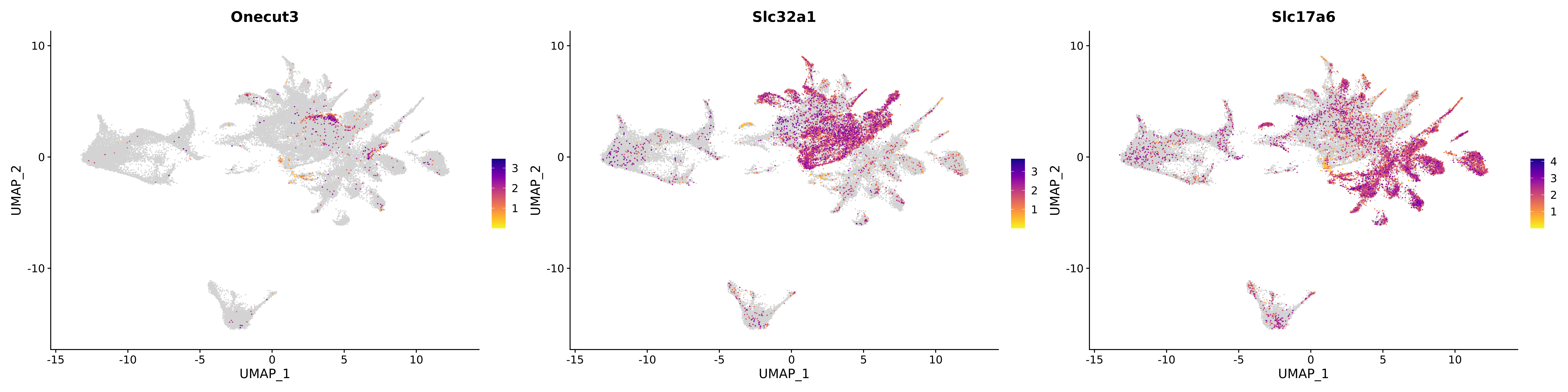
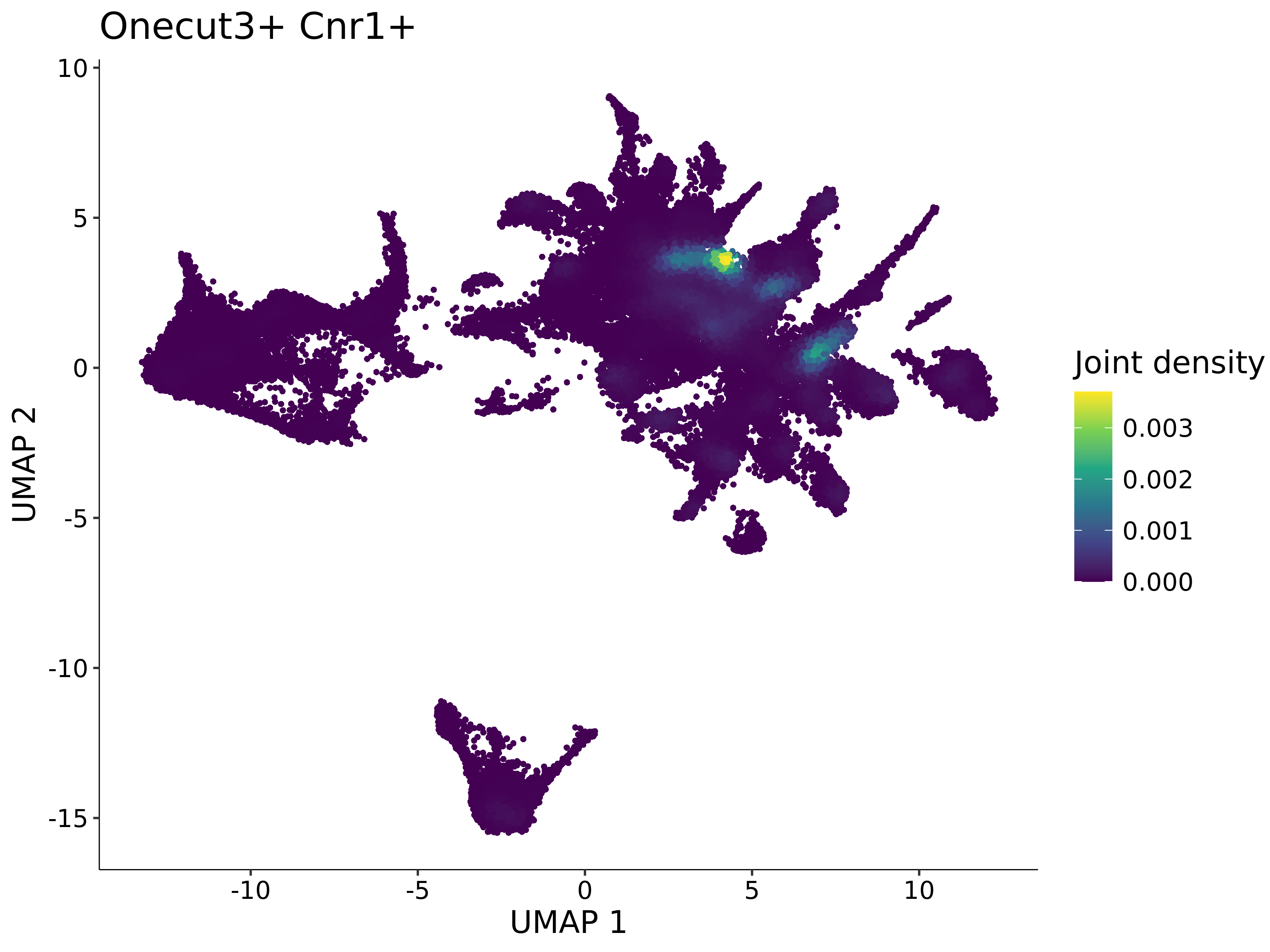
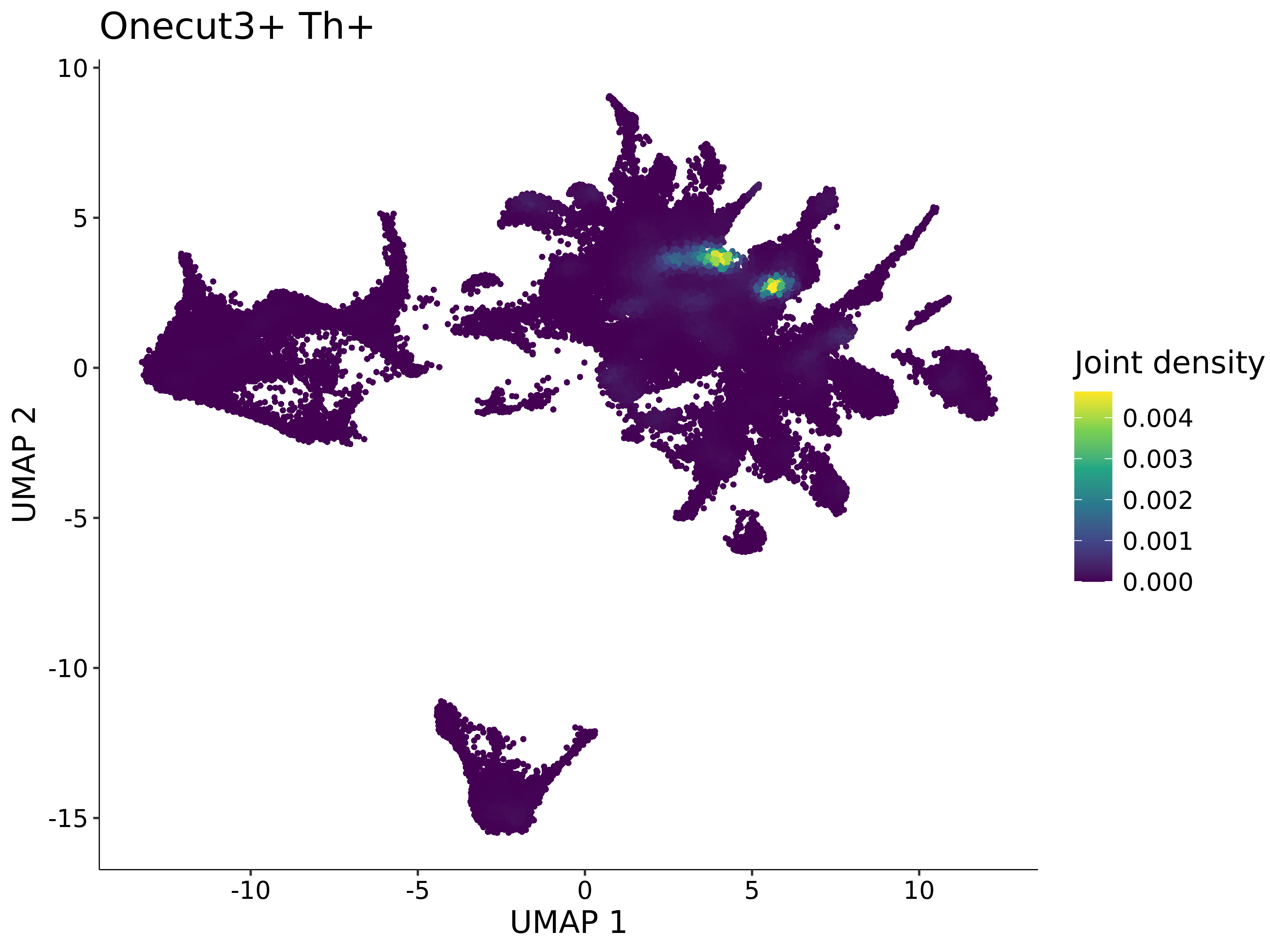
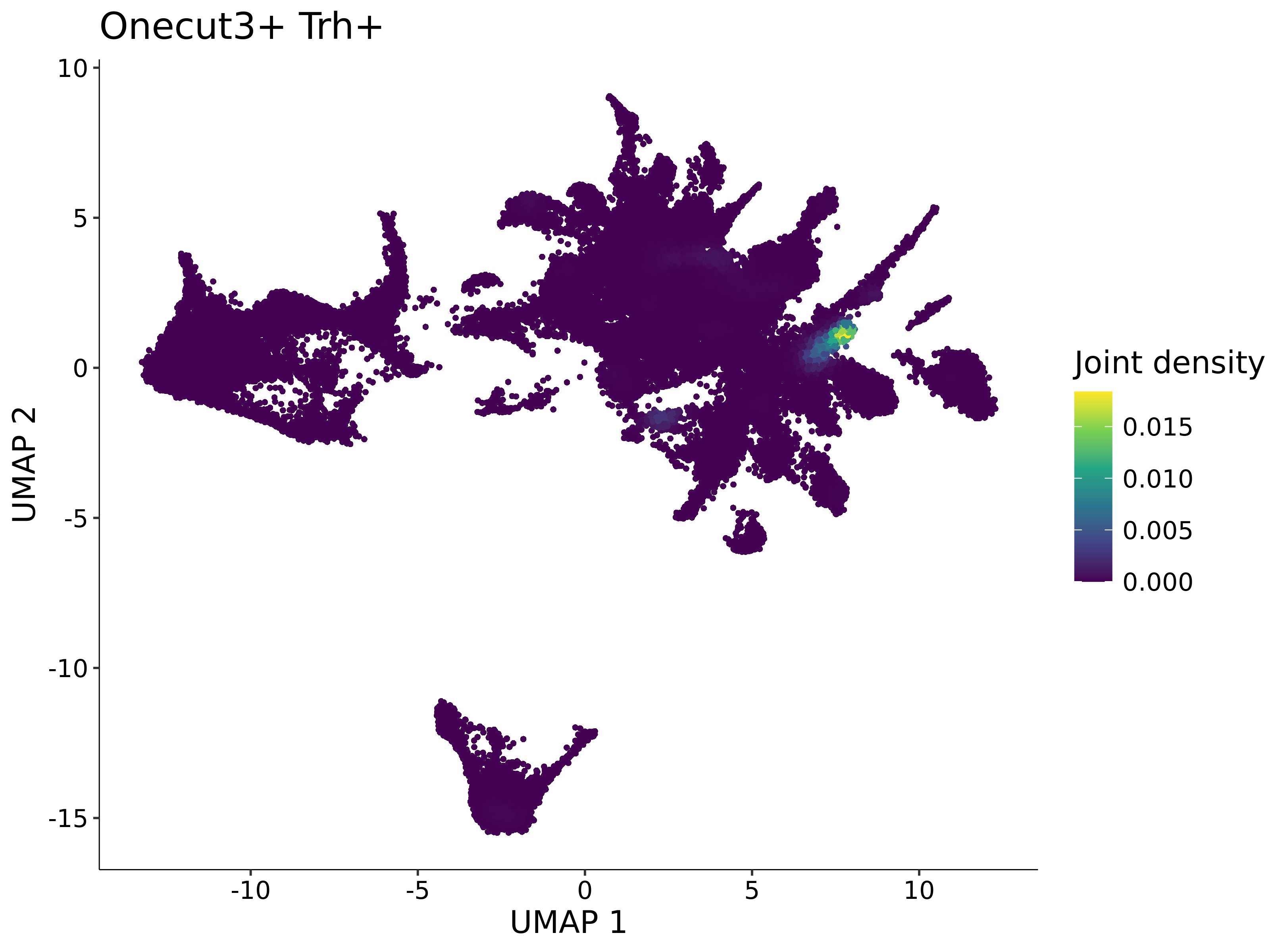
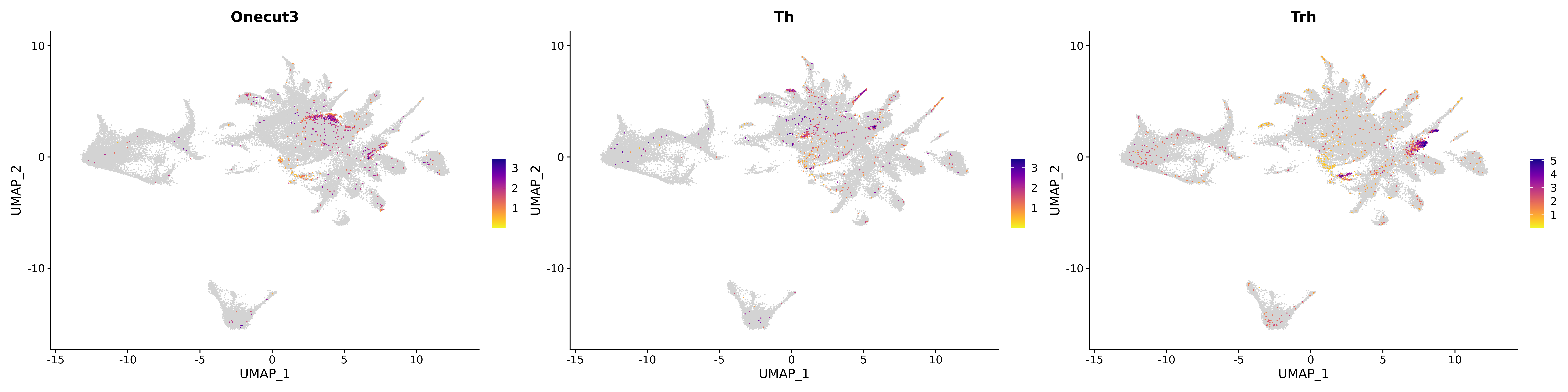
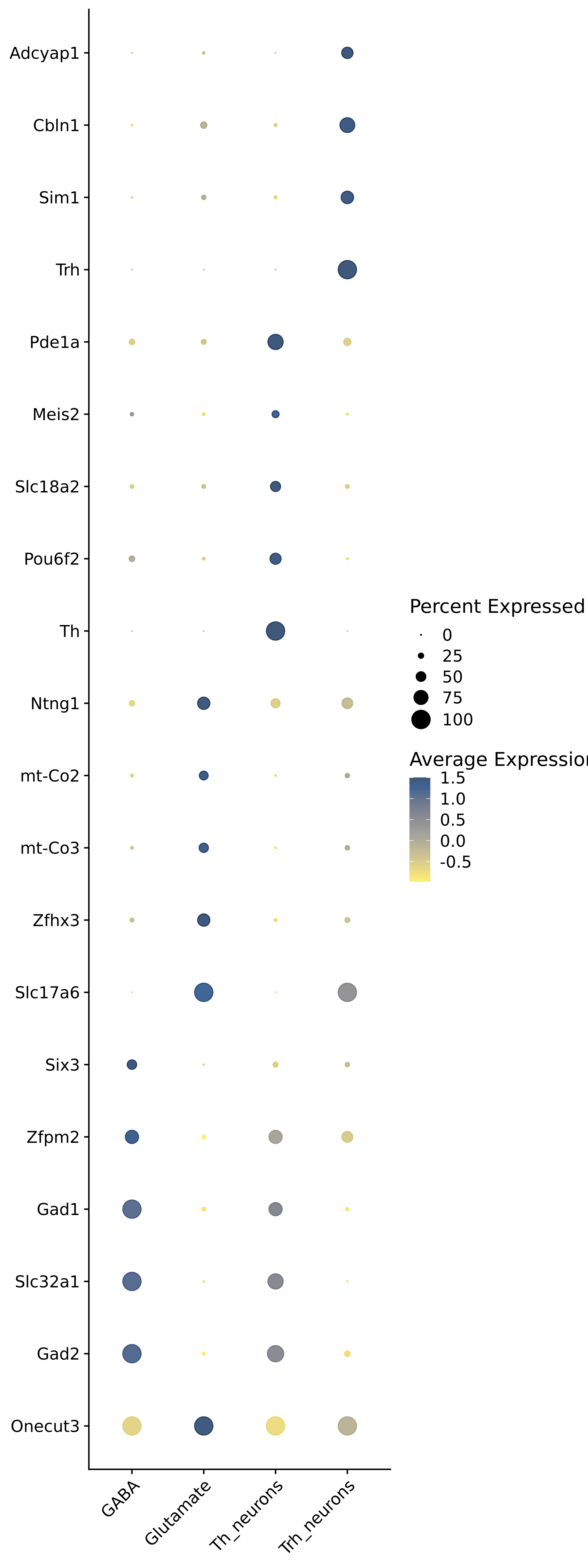
<- function (dat) {<- markOC3 %>% rownames_to_column (var = "gene" ) %>% filter (< 0.0000000001 ,str_detect (string = gene, pattern = "^mt-" , negate = T),str_detect (string = gene, pattern = "^Gm" , negate = T)%>% top_n (n = 40 , wt = avg_log2FC) %>% top_n (n = 20 , wt = pct.1 ) %>% :: arrange (desc (pct.1 )) %>% $ gene<- markOC3 %>% rownames_to_column (var = "gene" ) %>% filter (< 0.0000000001 ,str_detect (string = gene, pattern = "^mt-" , negate = T),str_detect (string = gene, pattern = "^Gm" , negate = T)%>% top_n (n = 25 , wt = p_val_adj) %>% top_n (n = 20 , wt = pct.1 ) %>% top_n (n = 15 , wt = avg_log2FC) %>% :: arrange (desc (pct.1 )) %>% $ gene<- GetAssayData (dat, "data" , "RNA" ) %>% as.data.frame () %>% t ()<- colSums (srt_onecut_filtered_ages) %>% > 10 ] %>% names () %>% %in% c ("Onecut1" ,"Onecut2" ,"Onecut3" %<>% .[, filt_age_oc_sum]<- corrr:: correlate (srt_onecut_filtered_ages)<- %>% arrange (- Onecut3) %>% select (term, Onecut3, Onecut2)<- .3 <- .3 <- .95 <- .95 <- intersect (x = oc3_age_corr_genes %>% slice_min (order_by = Onecut2,prop = threshold.min.pct.Onecut2y = oc3_age_corr_genes %>% slice_min (order_by = Onecut3,prop = threshold.max.pct.OC3<- union ($ term,<- c ("Onecut2" , "Pias2" , "Tp53" , "Trio" , "Fas" , "Rhoa" , "Limk1" , "Vegfa" , "Bcar1" ,"Ywhab" , "Nf1" , "Eif4e" , "Rb1" , "Sp1" , "Rps13" , "Rps3a" , "Traf6" , "Fgfr1op" ,"Ifi16" , "Irf3" , "Emg1" , "Myc" , "Pten" , "Sirt1" , "E2f1" , "Nr0b2" , "Crp" , "Erg" ,"Foxa1" , "Spi1" , "Nfkb1" , "Nfkbia" , "Pan2" , "Eif4a1" , "Gfra1" , "Stat6" , "Agt" ,"Onecut1" , "Grb2" ,$ gene_name %>% str_to_sentence ()%>% ! . %in% highlight_oc3_age_corr_genes] %>% unique () %>% %in% rownames (dat)]<- DotPlot (assay = "RNA" ,features = c (additional_genes, highlight_oc3_age_corr_genes),group.by = "age" + RotatedAxis ()save_plot (filename = paste0 (plots_dir, "/plot_dot_age_corrr_expression_wtin_OC23.pdf" ),plot = plot_dot_oc23_age_corr_genes,base_height = 6 ,base_aspect_ratio = 2 return (highlight_oc3_age_corr_genes)# oc_list <- Seurat::SplitObject(holder_srt$rar2020.srt.cont.oc2or3, split.by = "age")