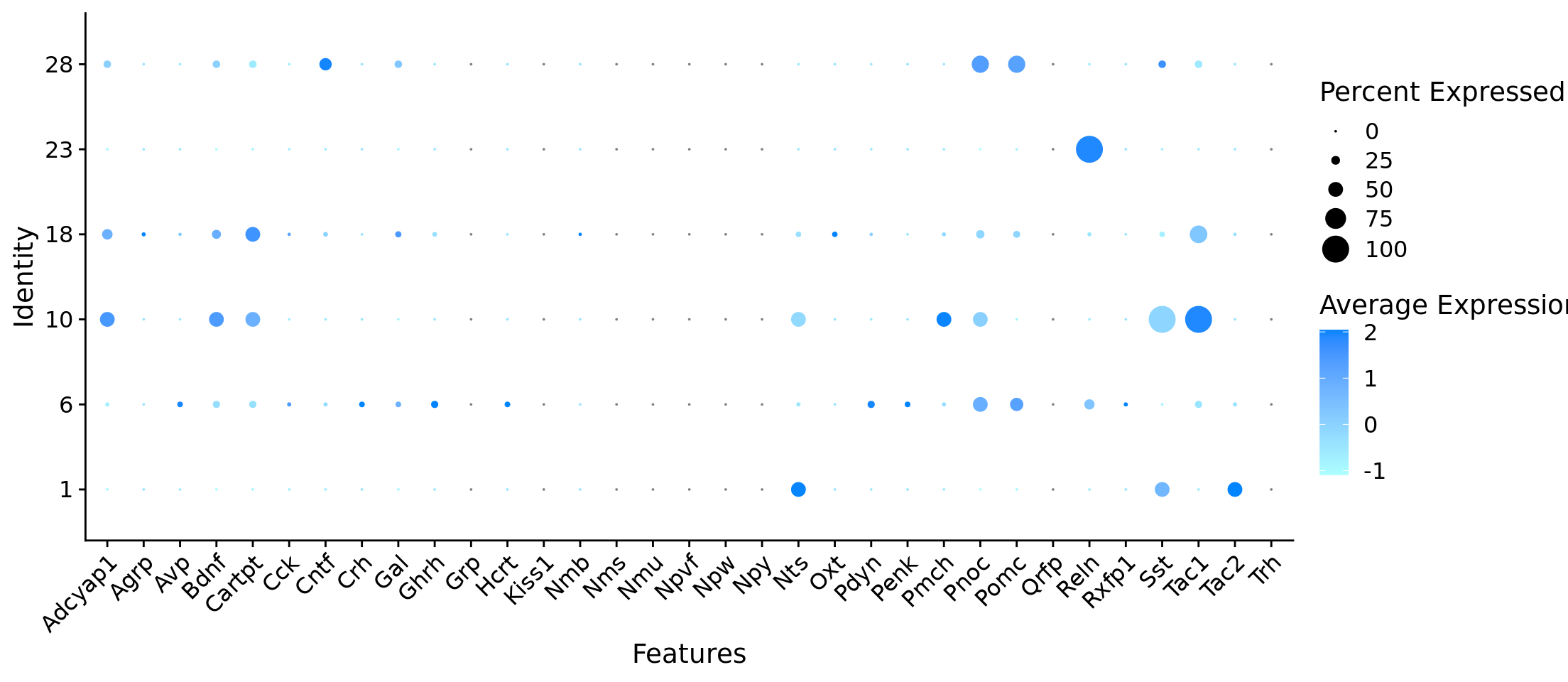
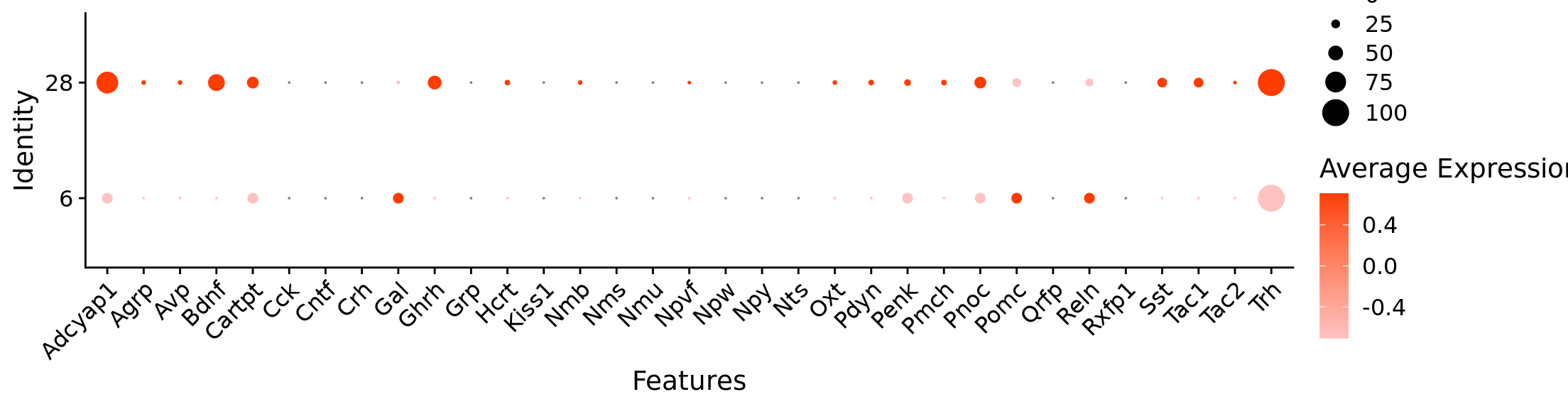
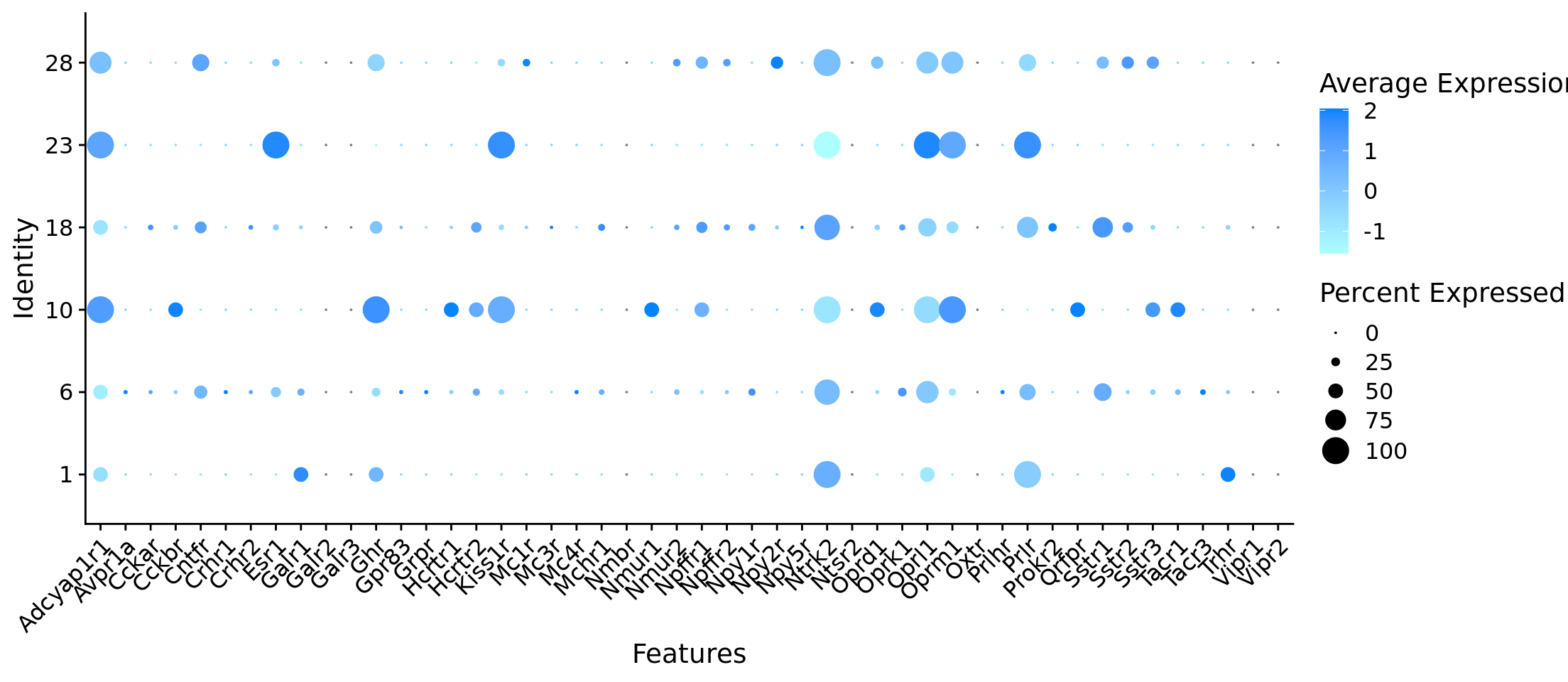
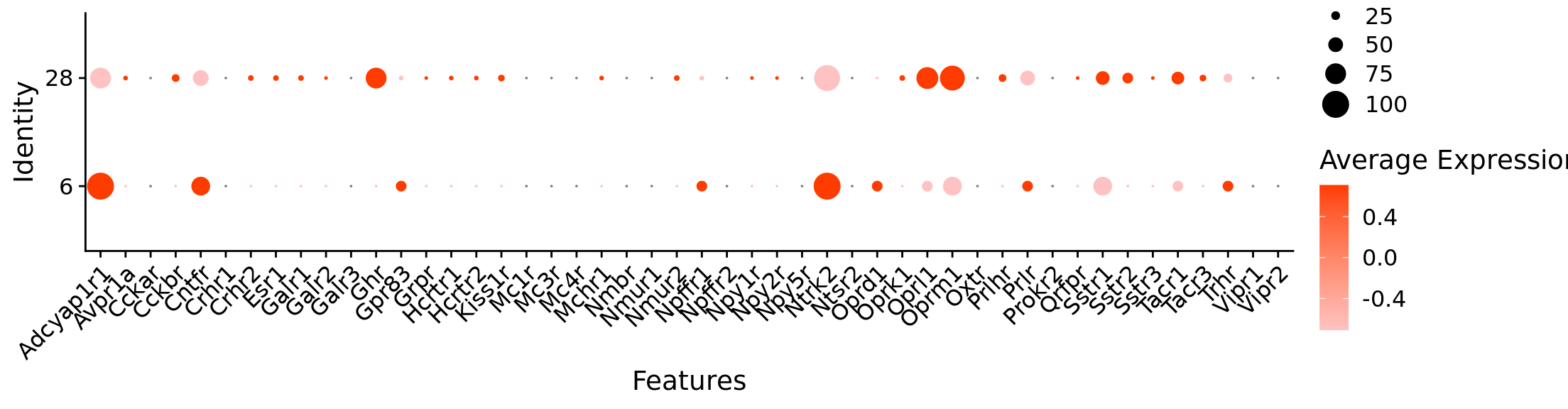
Rows: 384
Columns: 95
$ Slc17a6 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ Slc1a1 <dbl> 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0,…
$ Slc1a2 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ Slc1a6 <dbl> 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0,…
$ Gad1 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Slc32a1 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Slc6a1 <dbl> 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
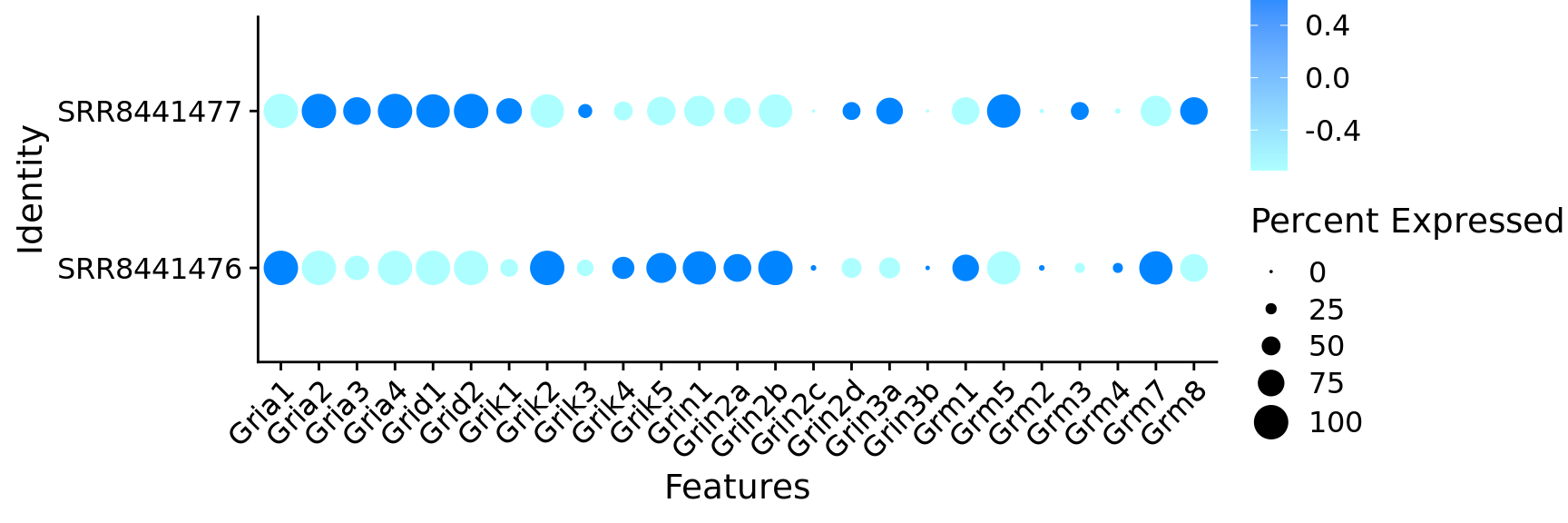
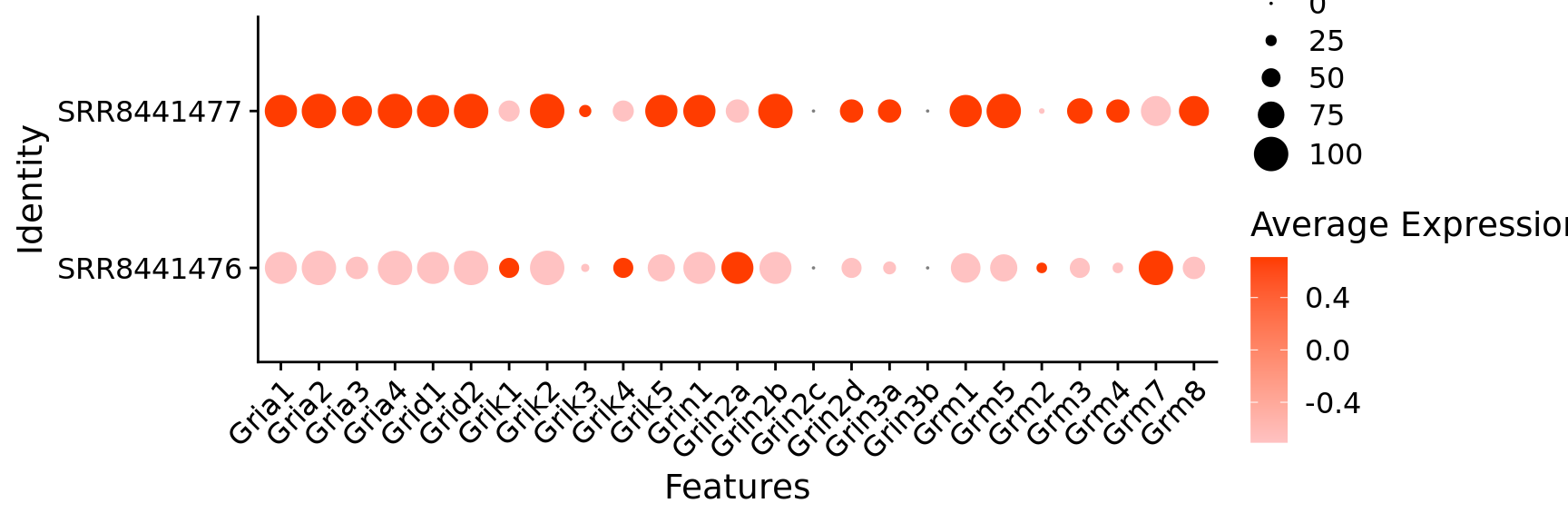
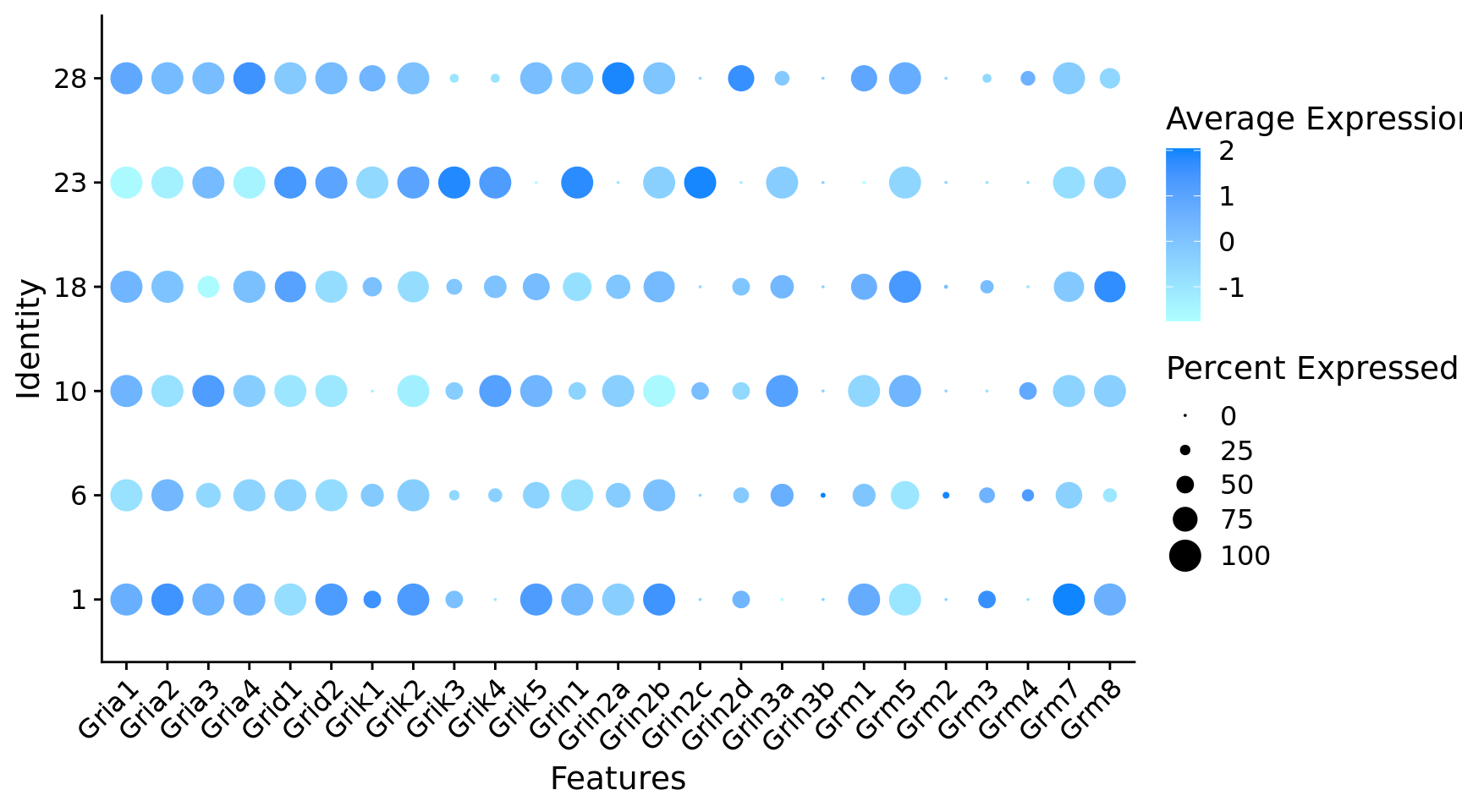
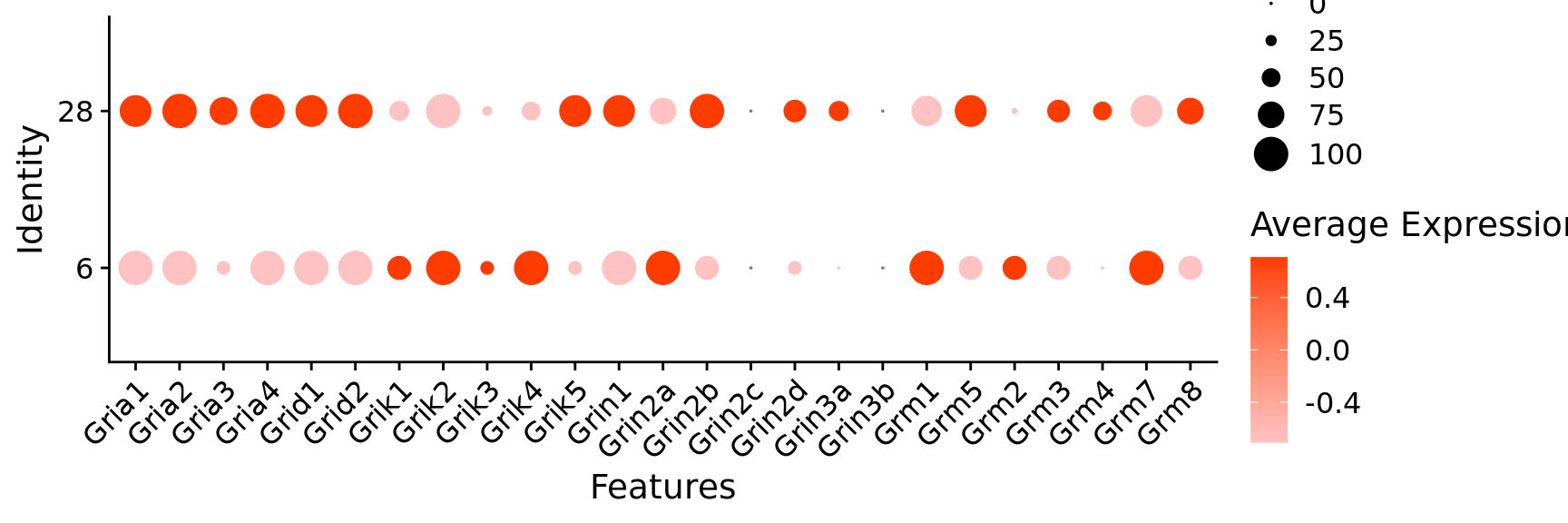
$ Gria1 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1,…
$ Gria2 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Gria3 <dbl> 1, 0, 1, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Gria4 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Grid1 <dbl> 1, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1,…
$ Grid2 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Grik1 <dbl> 1, 0, 1, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1, 1,…
$ Grik2 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Grik3 <dbl> 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 1, 1, 1, 0,…
$ Grik4 <dbl> 0, 0, 0, 1, 1, 1, 0, 1, 1, 0, 0, 0, 0, 0,…
$ Grik5 <dbl> 1, 1, 1, 0, 1, 1, 0, 1, 1, 0, 1, 0, 1, 1,…
$ Grin1 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0,…
$ Grin2a <dbl> 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0,…
$ Grin2b <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Grin2d <dbl> 1, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 1, 1, 1,…
$ Grin3a <dbl> 1, 0, 0, 0, 0, 1, 1, 1, 1, 1, 0, 1, 0, 1,…
$ Grm1 <dbl> 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1,…
$ Grm5 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0,…
$ Grm2 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ Grm3 <dbl> 0, 1, 1, 0, 0, 0, 1, 0, 1, 1, 0, 1, 1, 1,…
$ Grm4 <dbl> 0, 0, 0, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0,…
$ Grm7 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1,…
$ Grm8 <dbl> 1, 0, 1, 1, 0, 1, 0, 1, 0, 1, 0, 1, 1, 0,…
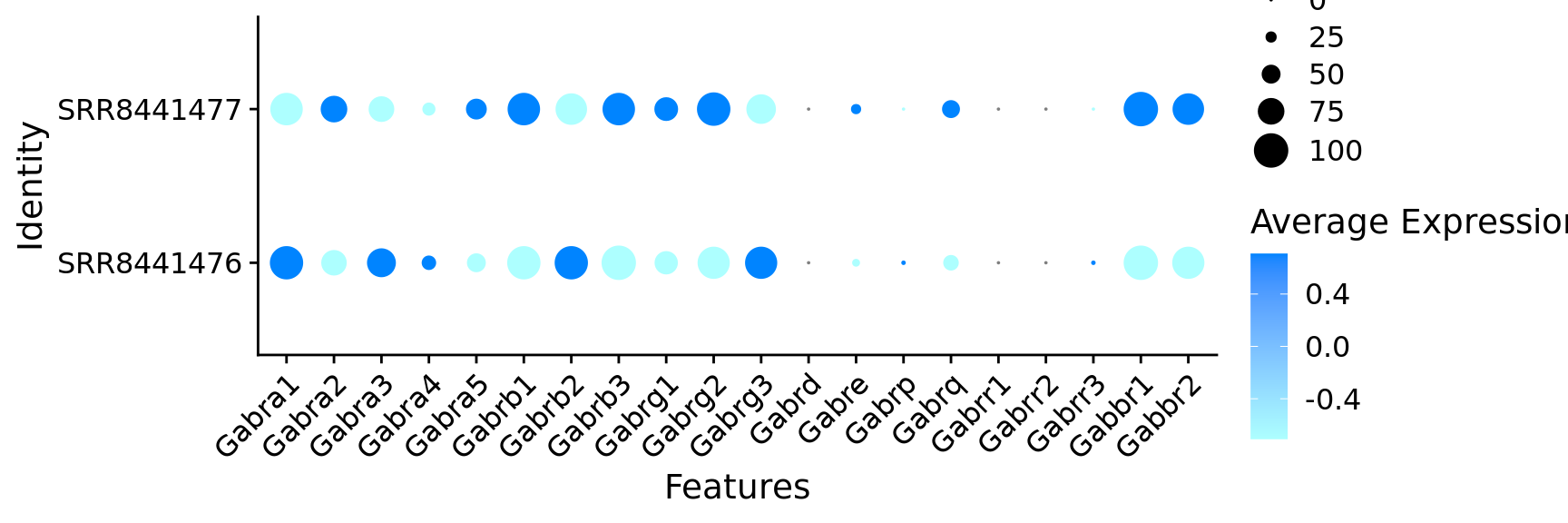
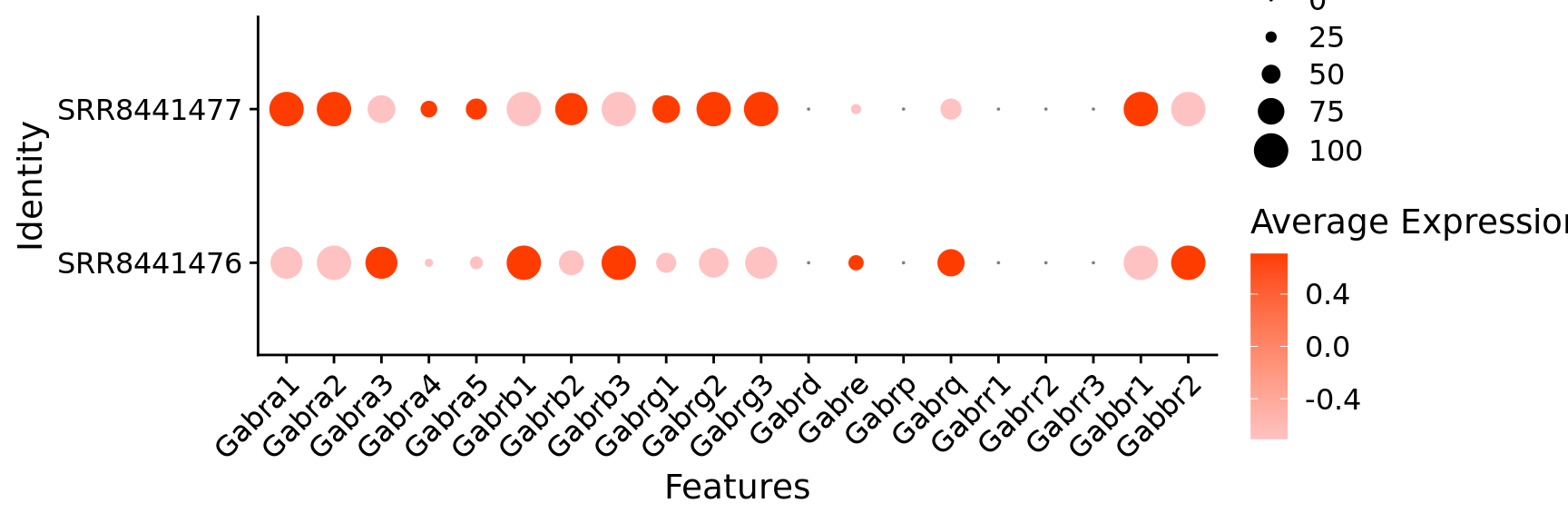
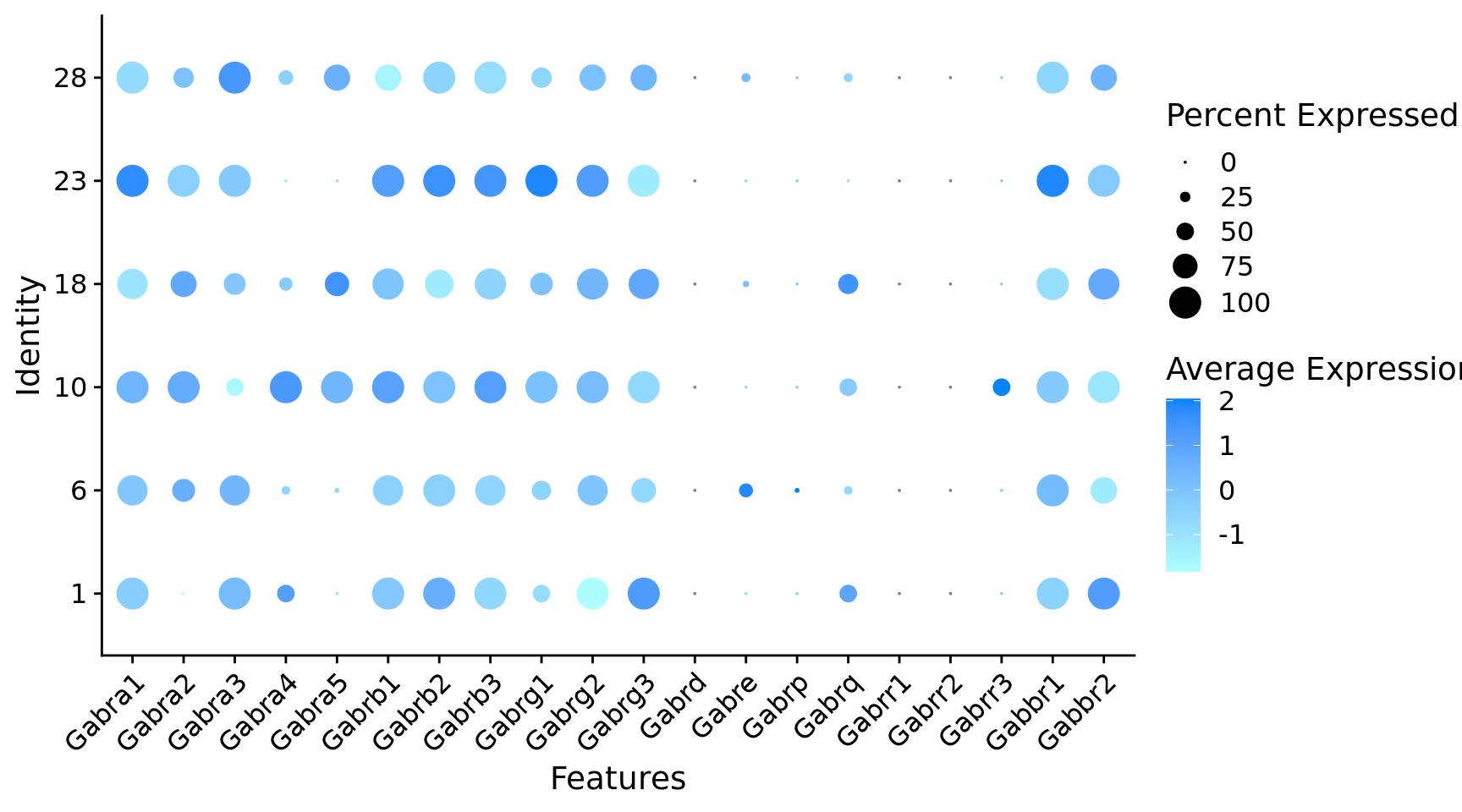
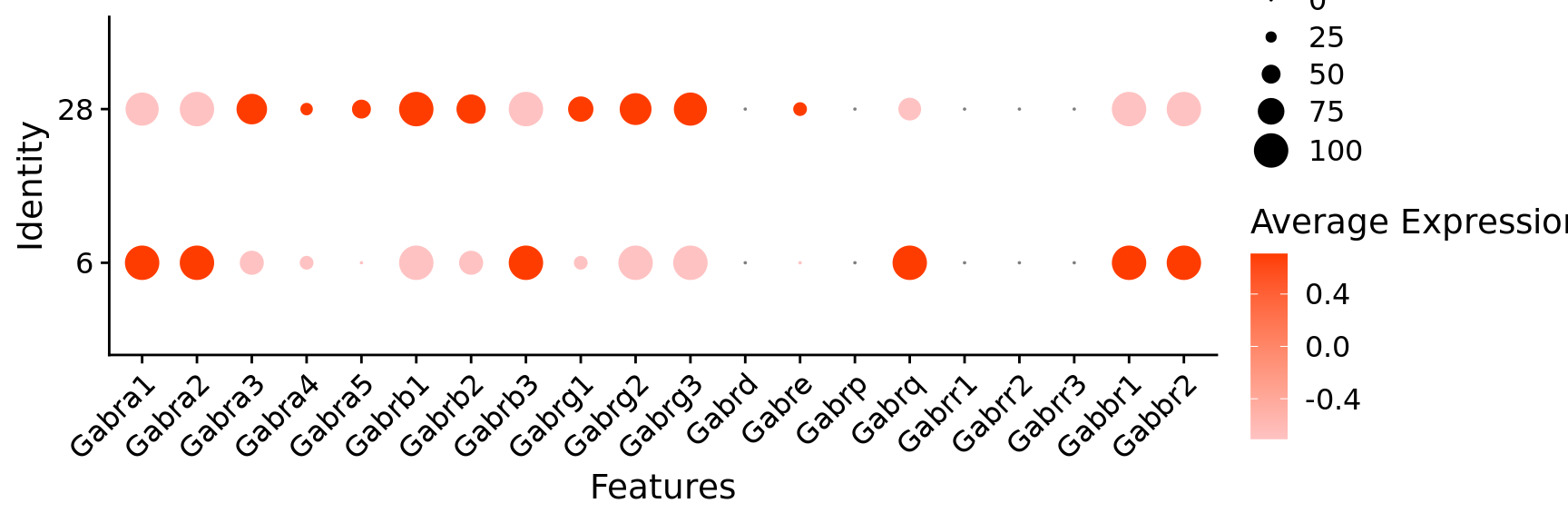
$ Gabra1 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1,…
$ Gabra2 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Gabra3 <dbl> 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 0, 0, 0, 0,…
$ Gabra4 <dbl> 0, 1, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0,…
$ Gabra5 <dbl> 1, 0, 0, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 1,…
$ Gabrb1 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Gabrb2 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Gabrb3 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Gabrg1 <dbl> 1, 1, 0, 1, 1, 0, 1, 0, 1, 1, 0, 0, 0, 0,…
$ Gabrg2 <dbl> 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Gabrg3 <dbl> 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
$ Gabre <dbl> 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0,…
$ Gabrq <dbl> 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0,…
$ Gabbr1 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0,…
$ Gabbr2 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1,…
$ Trh <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0,…
$ Th <dbl> 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0,…
$ Onecut3 <dbl> 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0,…
$ nCount_RAW <dbl> 19623, 12483, 12675, 13940, 15363, 15310,…
$ nFeature_RAW <int> 6025, 4802, 4842, 4980, 5118, 5211, 5297,…
$ nCount_RNA <dbl> 19162, 12185, 12373, 13618, 14957, 15002,…
$ nFeature_RNA <int> 5978, 4781, 4818, 4958, 5081, 5193, 5268,…
$ orig.ident <chr> "SRR8441476", "SRR8441476", "SRR8441476",…
$ nFeature_Diff <int> 47, 21, 24, 22, 37, 18, 29, 29, 31, 31, 1…
$ nCount_Diff <dbl> 461, 298, 302, 322, 406, 308, 376, 361, 3…
$ percent_mito <dbl> 4.075775, 3.947476, 2.327649, 5.742400, 1…
$ percent_ribo <dbl> 5.3908778, 8.1657776, 5.5524125, 6.865912…
$ percent_mito_ribo <dbl> 9.466653, 12.113254, 7.880061, 12.608313,…
$ percent_hb <dbl> 0.000000000, 0.000000000, 0.000000000, 0.…
$ log10GenesPerUMI <dbl> 0.8818700, 0.9005570, 0.8999119, 0.893857…
$ cell_name <chr> "SRR8441476_AAATGCCAGGCGACAT-1", "SRR8441…
$ barcode <chr> "AAATGCCAGGCGACAT-1", "AACCGCGAGCGGATCA-1…
$ latent_RT_efficiency <dbl> 3.283587, 2.502992, 2.562110, 2.646928, 2…
$ latent_cell_probability <dbl> 0.9999477, 0.9998801, 0.9999301, 0.999928…
$ latent_scale <dbl> 4695.590, 4008.599, 3921.173, 4068.829, 4…
$ doublet_score <dbl> 0.04804282, 0.07987788, 0.05905781, 0.068…
$ predicted_doublets <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ QC <chr> "Pass", "Pass", "Pass", "Pass", "Pass", "…
$ var_regex <dbl> 10.766100, 13.254001, 9.140871, 14.003525…
$ RNA_snn_res.0.5 <fct> 4, 5, 11, 5, 4, 5, 4, 4, 4, 4, 4, 4, 4, 1…
$ RNA_snn_res.0.74082856775745 <fct> 3, 6, 13, 6, 3, 9, 3, 3, 3, 3, 3, 3, 3, 4…
$ RNA_snn_res.1.24733796060143 <fct> 5, 6, 16, 6, 5, 11, 5, 9, 9, 28, 28, 9, 9…
$ RNA_snn_res.3.000001 <fct> 11, 13, 17, 13, 11, 12, 11, 8, 8, 27, 27,…
$ seurat_clusters <fct> 3, 20, 27, 20, 30, 19, 10, 3, 3, 10, 11, …
$ k_tree <fct> 9, 5, 23, 5, 1, 14, 1, 9, 9, 1, 1, 9, 4, …
$ comb_clstr1 <fct> 3, 6, 13, 6, 3, 9, 3, 3, 3, 3, 3, 3, 3, 4…
$ S.Score <dbl> 0.001711877, -0.019196776, -0.038780572, …
$ G2M.Score <dbl> 0.022455905, 0.001625043, -0.014130781, -…
$ Phase <chr> "G2M", "G2M", "G1", "S", "G2M", "G1", "G2…
$ nCount_SCT <dbl> 5786, 6661, 6617, 6344, 6031, 6118, 6219,…
$ nFeature_SCT <int> 3074, 3904, 3858, 3495, 3117, 3251, 3442,…
$ SCT_snn_res.1 <fct> 9, 5, 20, 5, 4, 13, 4, 9, 9, 4, 4, 9, 4, …
$ SCT_snn_res.1.10569909553377 <fct> 9, 5, 20, 5, 4, 12, 4, 9, 9, 4, 4, 9, 6, …
$ SCT_snn_res.1.23112793047964 <fct> 9, 5, 22, 5, 2, 12, 2, 9, 9, 2, 2, 9, 6, …
$ SCT_snn_res.1.38237925427927 <fct> 9, 5, 23, 5, 2, 12, 2, 9, 9, 2, 2, 9, 6, …
$ SCT_snn_res.1.5683423560766 <fct> 9, 5, 22, 5, 2, 12, 2, 9, 9, 2, 2, 9, 6, …
$ SCT_snn_res.1.80251553997986 <fct> 9, 5, 23, 5, 1, 14, 1, 9, 9, 1, 1, 9, 4, …
$ SCT_snn_res.2.10643815972253 <fct> 7, 19, 26, 19, 2, 13, 2, 7, 7, 2, 2, 7, 4…
$ SCT_snn_res.2.51672603000175 <fct> 6, 20, 26, 20, 4, 14, 4, 6, 6, 4, 7, 6, 7…
$ SCT_snn_res.3.10106021549219 <fct> 6, 20, 26, 20, 5, 12, 5, 6, 6, 9, 9, 6, 9…
$ SCT_snn_res.4.00000100000001 <fct> 3, 20, 27, 20, 30, 19, 10, 3, 3, 10, 11, …
$ gaba_status <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,…
$ gaba_occurs <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,…
$ glut_status <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,…
$ status <chr> "query", "query", "query", "query", "quer…