Load data and setup parameters
Code
# Load tidyverse infrastructure packages library (here)library (tidyverse)library (RColorBrewer)library (glmnet)# Load packages for scRNA-seq analysis and visualisation library (Seurat)library (SeuratWrappers)library (SeuratDisk)library (scCustomize)library (swne)library (ggplot2)library (cowplot)library (UpSetR)library (patchwork)library (Nebulosa)library (schex)
Set paths
Code
set_here ()<- here ("data" )<- here ("outputs" )<- here (output_dir, "figures" )<- here (output_dir, "tables" )
Load helper functions and gene-sets
Code
source (here ("functions.R" ))source (here ("genes.R" ))
Set fixed variables
Code
# set seed <- 42 set.seed (seed = reseed)# available RAM in kB <- check_ram ()# available cores <- available_cores (prop2use = .1 )# Parameters for parallel execution plan ("multisession" , workers = n.cores)options (future.globals.maxSize = 100000 * 1024 ^ 2 ,future.rng.onMisuse = "ignore" plan ()
multisession:
- args: function (..., workers = 8, envir = parent.frame())
- tweaked: TRUE
- call: plan("multisession", workers = n.cores)
Load data from Zeisel et al. (2018)
Code
<- Connect (filename = here ("l6_r1_neurons.loom" ), mode = "r" )
Class: loom
Filename: /data/l6_r1_neurons.loom
Access type: H5F_ACC_RDONLY
Attributes: last_modified
Listing:
name obj_type dataset.dims dataset.type_class
attrs H5I_GROUP <NA> <NA>
col_attrs H5I_GROUP <NA> <NA>
col_graphs H5I_GROUP <NA> <NA>
layers H5I_GROUP <NA> <NA>
matrix H5I_DATASET 74539 x 27998 H5T_FLOAT
row_attrs H5I_GROUP <NA> <NA>
row_graphs H5I_GROUP <NA> <NA>
Code
<- as.Seurat (l6.neurons)$ close_all ()
show srt object
Code
<- Store_Palette_Seurat (seurat_object = l6.srt,palette = rev (brewer.pal (n = 11 , name = "Spectral" )),palette_name = "div_Colour_Pal"
An object of class Seurat
27998 features across 74539 samples within 1 assay
Active assay: RNA (27998 features, 0 variable features)
show metadata
Code
:: skim (l6.srt@ meta.data)
Data summary
Name
l6.srt@meta.data
Number of rows
74539
Number of columns
129
_______________________
Column type frequency:
character
78
factor
1
numeric
50
________________________
Group variables
None
Variable type: character
Age
0
1
1
8
0
22
0
AnalysisPool
0
1
0
15
48185
12
0
AnalysisProject
0
1
0
10
48185
2
0
Bucket
0
1
47
65
0
19
0
CellConc
0
1
0
4
48185
7
0
Cell_Conc
0
1
0
4
48185
8
0
ChipID
0
1
0
5
48185
23
0
Class
0
1
7
7
0
1
0
ClusterName
0
1
2
7
0
214
0
Comment
0
1
3
81
0
55
0
Comments
0
1
0
168
48185
30
0
DateCaptured
0
1
0
19
48185
22
0
Date_Captured
0
1
0
10
48185
22
0
Description
0
1
14
68
0
93
0
Developmental_compartment
0
1
11
15
0
6
0
DonorID
0
1
0
25
48185
24
0
Estimated.Number.of.Cells
0
1
0
5
70736
13
0
Flowcell
0
1
0
19
48185
20
0
Fraction.Reads.in.Cells
0
1
0
5
48185
61
0
Label
0
1
0
9
73578
6
0
Location_based_on
0
1
0
40
11871
175
0
Mean.Reads.per.Cell
0
1
0
7
48185
73
0
Median.Genes.per.Cell
0
1
0
5
48185
72
0
Median.UMI.Counts.per.Cell
0
1
0
6
69156
15
0
NGI_PlateWell
0
1
0
0
74539
1
0
Neurotransmitter
0
1
0
47
2875
38
0
NumPooledAnimals
0
1
0
3
48185
4
0
Num_Pooled_Animals
0
1
0
1
48185
4
0
Number.of.Reads
0
1
0
11
48185
73
0
PCRCycles
0
1
0
4
48185
4
0
PCR_Cycles
0
1
0
2
48185
4
0
PassedQC
0
1
0
2
48185
2
0
PlugDate
0
1
0
3
48185
2
0
Plug_Date
0
1
0
0
74539
1
0
Probable_location
0
1
3
128
0
133
0
Project
0
1
0
10
48185
3
0
Q30.Bases.in.Barcode
0
1
0
5
48185
53
0
Q30.Bases.in.RNA.Read
0
1
0
5
70736
13
0
Q30.Bases.in.Sample.Index
0
1
0
5
70736
12
0
Q30.Bases.in.UMI
0
1
0
5
48185
51
0
Reads.Mapped.Confidently.to.Exonic.Regions
0
1
0
5
48185
67
0
Reads.Mapped.Confidently.to.Intergenic.Regions
0
1
0
5
48185
35
0
Reads.Mapped.Confidently.to.Intronic.Regions
0
1
0
5
48185
62
0
Reads.Mapped.Confidently.to.Transcriptome
0
1
0
5
48185
68
0
Region
0
1
4
48
0
25
0
SampleID
0
1
7
7
0
131
0
SampleIndex
0
1
0
9
48185
67
0
SampleOK
0
1
0
2
73468
2
0
Sample_Index
0
1
0
9
59706
31
0
SeqComment
0
1
0
3
48185
2
0
SeqLibDate
0
1
0
19
48185
4
0
SeqLibOk
0
1
0
3
48185
3
0
Seq_Comment
0
1
0
0
74539
1
0
Seq_Lib_Date
0
1
0
9
73661
3
0
Seq_Lib_Ok
0
1
0
1
73661
2
0
Sequencing.Saturation
0
1
0
5
70736
13
0
Serial_Number
0
1
0
3
48185
73
0
Sex
0
1
1
5
0
12
0
Species
0
1
0
2
48185
2
0
Strain
0
1
0
27
48185
5
0
Subclass
0
1
7
15
0
2
0
TargetNumCells
0
1
0
5
48185
8
0
Target_Num_Cells
0
1
0
5
48185
9
0
TaxonomyRank1
0
1
7
7
0
1
0
TaxonomyRank2
0
1
11
11
0
2
0
TaxonomyRank3
0
1
15
50
0
11
0
TaxonomyRank4
0
1
15
43
0
21
0
TaxonomySymbol
0
1
4
4
0
21
0
Taxonomy_group
0
1
15
43
0
21
0
TimepointPool
0
1
0
3
48185
2
0
Tissue
0
1
2
10
0
23
0
Total.Genes.Detected
0
1
0
6
70736
13
0
Transcriptome
0
1
0
4
48185
2
0
Valid.Barcodes
0
1
0
5
48185
41
0
cDNAConcNanogramPerMicroliter
0
1
0
18
48185
56
0
cDNALibOk
0
1
0
9
48185
4
0
cDNA_Lib_Ok
0
1
0
9
54253
3
0
ngperul_cDNA
0
1
0
4
48185
56
0
Variable type: factor
orig.ident
0
1
FALSE
35
10X: 7100, 10X: 6905, 10X: 6766, 10X: 6515
Variable type: numeric
nCount_RNA
0
1
4184.82
3644.92
160.00
1805.00
3199.00
5396.00
73358.00
▇▁▁▁▁
nFeature_RNA
0
1
1926.62
1029.78
88.00
1109.00
1776.00
2597.00
7490.00
▇▇▂▁▁
ClassProbability_Astrocyte
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.28
▇▁▁▁▁
ClassProbability_Astrocyte.Immune
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.22
▇▁▁▁▁
ClassProbability_Astrocyte.Neurons
0
1
0.00
0.01
0.00
0.00
0.00
0.00
0.35
▇▁▁▁▁
ClassProbability_Astrocyte.Oligos
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.02
▇▁▁▁▁
ClassProbability_Astrocyte.Vascular
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.04
▇▁▁▁▁
ClassProbability_Bergmann.glia
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.22
▇▁▁▁▁
ClassProbability_Blood
0
1
0.00
0.01
0.00
0.00
0.00
0.00
0.36
▇▁▁▁▁
ClassProbability_Blood.Vascular
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.46
▇▁▁▁▁
ClassProbability_Enteric.glia
0
1
0.00
0.01
0.00
0.00
0.00
0.00
0.80
▇▁▁▁▁
ClassProbability_Enteric.glia.Cycling
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.01
▇▁▁▁▁
ClassProbability_Ependymal
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.40
▇▁▁▁▁
ClassProbability_Ex.Neurons
0
1
0.00
0.00
0.00
0.00
0.00
0.01
0.09
▇▁▁▁▁
ClassProbability_Ex.Vascular
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.11
▇▁▁▁▁
ClassProbability_Immune
0
1
0.00
0.01
0.00
0.00
0.00
0.00
0.40
▇▁▁▁▁
ClassProbability_Immune.Neurons
0
1
0.00
0.01
0.00
0.00
0.00
0.00
0.55
▇▁▁▁▁
ClassProbability_Immune.Oligos
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.01
▇▁▁▁▁
ClassProbability_Neurons
0
1
0.97
0.10
0.00
0.98
0.99
0.99
1.00
▁▁▁▁▇
ClassProbability_Neurons.Cycling
0
1
0.01
0.10
0.00
0.00
0.00
0.00
1.00
▇▁▁▁▁
ClassProbability_Neurons.Oligos
0
1
0.01
0.02
0.00
0.00
0.00
0.01
0.49
▇▁▁▁▁
ClassProbability_Neurons.Satellite.glia
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.39
▇▁▁▁▁
ClassProbability_Neurons.Vascular
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.27
▇▁▁▁▁
ClassProbability_OEC
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.33
▇▁▁▁▁
ClassProbability_Oligos
0
1
0.00
0.01
0.00
0.00
0.00
0.00
0.45
▇▁▁▁▁
ClassProbability_Oligos.Cycling
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.28
▇▁▁▁▁
ClassProbability_Oligos.Vascular
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.02
▇▁▁▁▁
ClassProbability_Satellite.glia
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.01
▇▁▁▁▁
ClassProbability_Satellite.glia.Cycling
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.01
▇▁▁▁▁
ClassProbability_Satellite.glia.Schwann
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.00
▇▁▁▁▁
ClassProbability_Schwann
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.00
▇▁▁▁▁
ClassProbability_Ttr
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.30
▇▁▁▁▁
ClassProbability_Vascular
0
1
0.00
0.01
0.00
0.00
0.00
0.00
0.33
▇▁▁▁▁
Clusters
0
1
108.63
52.78
0.00
71.00
94.00
160.00
213.00
▃▇▆▇▃
LeafOrder
0
1
70.80
61.76
1.00
24.00
42.00
128.00
214.00
▇▃▁▂▂
MitoRiboRatio
0
1
0.44
1.54
0.00
0.14
0.23
0.37
132.75
▇▁▁▁▁
OriginalClusters
0
1
7.55
6.40
0.00
2.00
6.00
12.00
24.00
▇▅▂▃▁
Outliers
0
1
0.00
0.00
0.00
0.00
0.00
0.00
0.00
▁▁▇▁▁
X_KMeans_10
0
1
0.96
1.74
0.00
0.00
0.00
1.00
10.00
▇▁▁▁▁
X_LogCV
0
1
1.27
1.77
0.00
0.00
0.00
3.06
6.27
▇▁▂▂▁
X_LogMean
0
1
-1.22
1.82
-6.33
-2.80
0.00
0.00
1.39
▁▂▂▁▇
X_NGenes
0
1
1926.62
1029.78
88.00
1109.00
1776.00
2597.00
7490.00
▇▇▂▁▁
X_PC1
0
1
0.27
6.90
-81.88
0.00
0.00
0.00
108.10
▁▁▇▁▁
X_PC2
0
1
0.00
0.41
-13.24
0.00
0.00
0.00
12.79
▁▁▇▁▁
X_Total
0
1
4184.82
3644.92
160.00
1805.00
3199.00
5396.00
73358.00
▇▁▁▁▁
X_Valid
0
1
1.00
0.00
1.00
1.00
1.00
1.00
1.00
▁▁▇▁▁
X_X
0
1
0.00
15.34
-32.75
-12.21
-0.39
12.18
33.68
▃▆▇▇▃
X_Y
0
1
0.00
15.08
-34.33
-10.69
0.47
11.77
32.94
▂▆▇▆▂
X_tSNE1
0
1
0.11
12.08
-53.06
0.00
0.00
0.00
54.05
▁▁▇▁▁
X_tSNE2
0
1
1.59
11.60
-51.77
0.00
0.00
0.00
52.87
▁▁▇▁▁
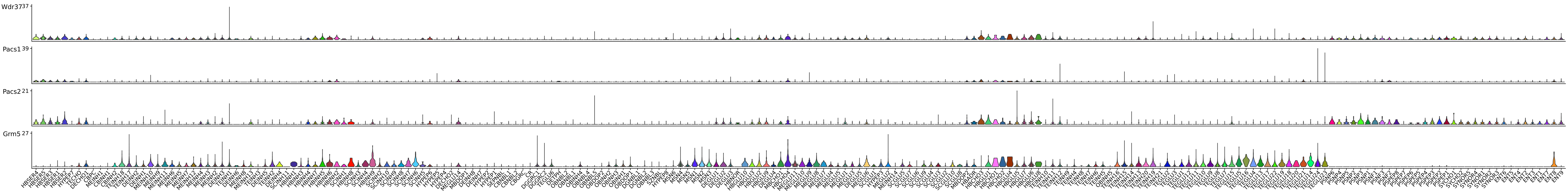
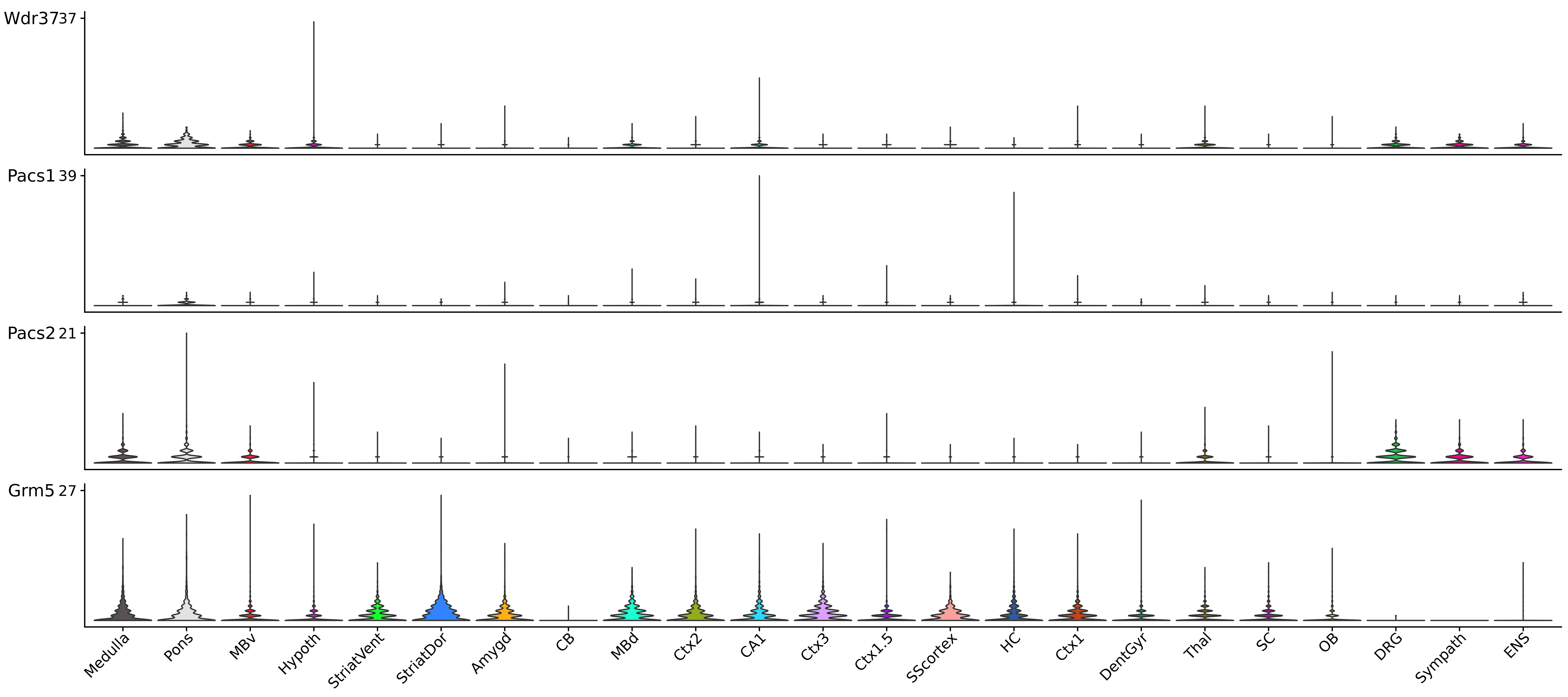
Check Violin plots for the expression of the gene of interest (goi) in the different clusters and tissues
Code
Idents (l6.srt) <- "ClusterName" Stacked_VlnPlot (seurat_object = l6.srt, features = goi, x_lab_rotate = TRUE ,color_seed = reseed
Code
Idents (l6.srt) <- "Tissue" Stacked_VlnPlot (seurat_object = l6.srt, features = goi, x_lab_rotate = TRUE ,color_seed = reseed
Code
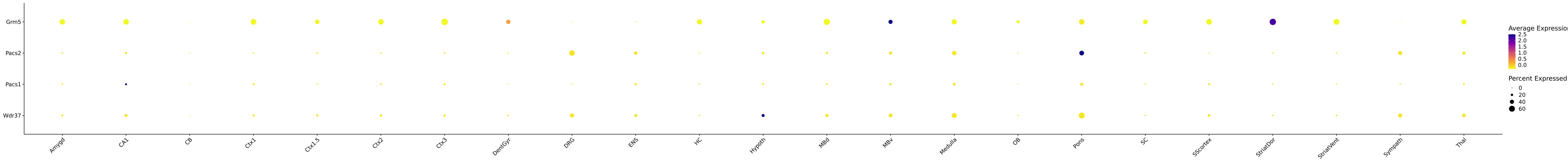
DotPlot_scCustom (seurat_object = l6.srt,assay = "RNA" ,features = goi,flip_axes = TRUE ,x_lab_rotate = TRUE ,group.by = "Tissue"
Code
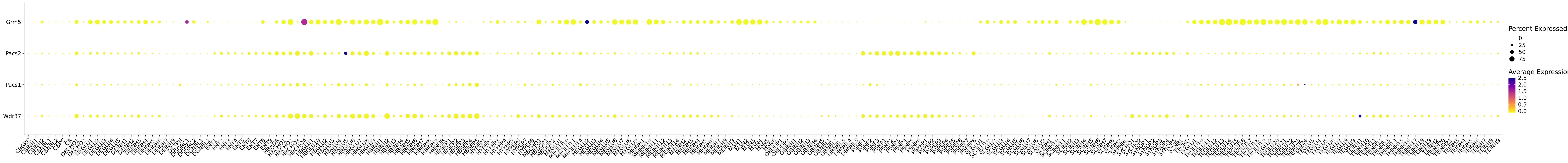
DotPlot_scCustom (seurat_object = l6.srt,assay = "RNA" ,features = goi,flip_axes = TRUE ,x_lab_rotate = TRUE ,group.by = "ClusterName"
Extact UMAP and tSNE coordinates from the srt object
Code
<- l6.srt@ meta.data %>% select (X_X, X_Y) %>% as.matrix ()rownames (umap) <- colnames (l6.srt)colnames (umap) <- paste0 ("UMAP_" , 1 : 2 )"umap" ]] <- CreateDimReducObject (embeddings = umap, key = "UMAP_" , assay = DefaultAssay (l6.srt))<- l6.srt@ meta.data %>% select (X_tSNE1, X_tSNE2) %>% as.matrix ()rownames (tsne) <- colnames (l6.srt)colnames (tsne) <- paste0 ("tSNE_" , 1 : 2 )"tsne" ]] <- CreateDimReducObject (embeddings = tsne, key = "tSNE_" , assay = DefaultAssay (l6.srt))
Exploratory data analysis
Use standradised pipeline
Code
<- NormalizeData (l6.srt)<- FindVariableFeatures (l6.srt, selection.method = "vst" , nfeatures = 3500 )<- ScaleData (l6.srt)<- VariableFeatures (l6.srt)<- Idents (l6.srt)
Check UMAP and tSNE plots for the expression of the gene of interest (goi) in the different clusters and tissues
tSNE
Code
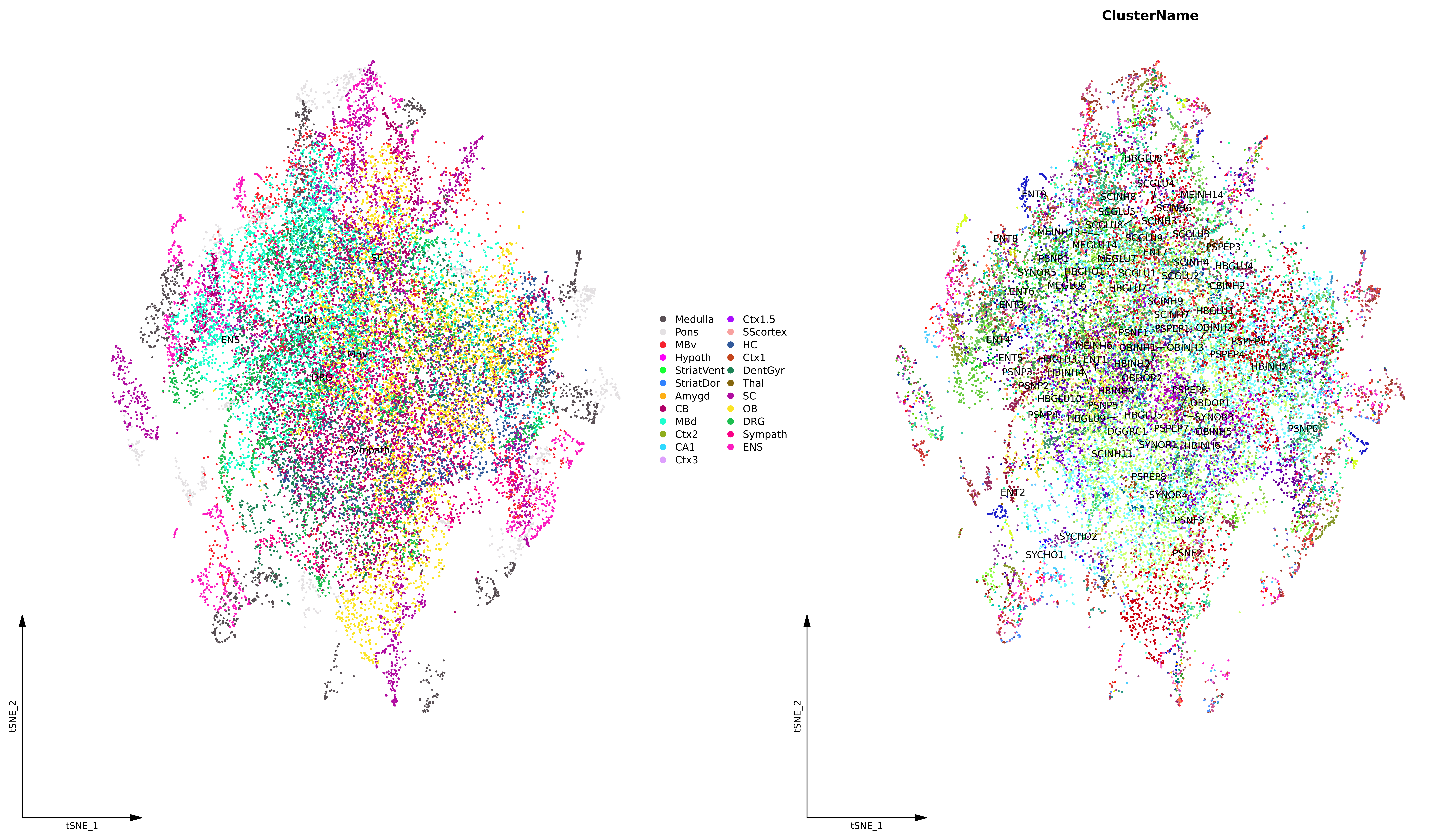
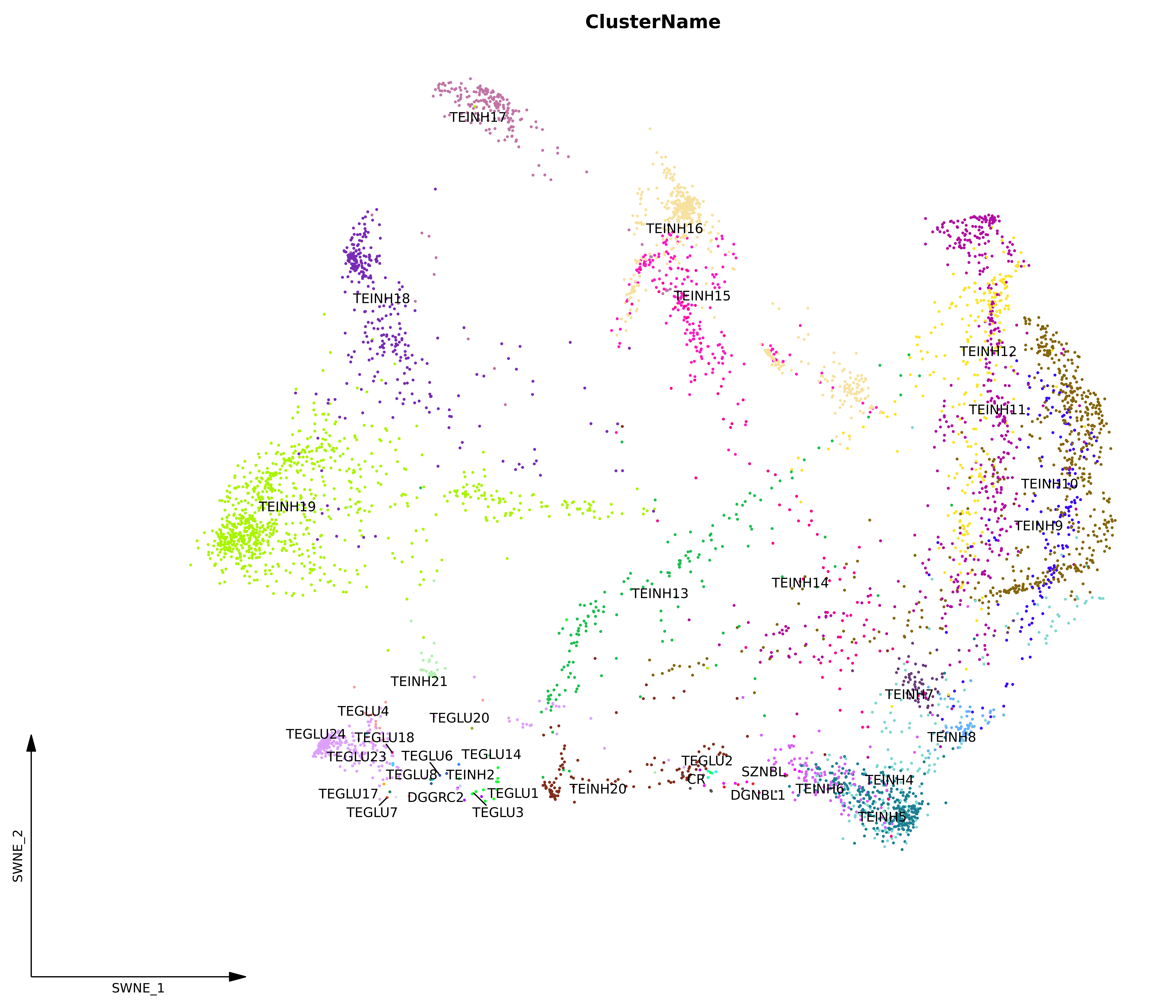
DimPlot_scCustom (reduction = "tsne" ,pt.size = 0.5 ,label = TRUE ,repel = TRUE ,figure_plot = TRUE ,color_seed = reseed| DimPlot_scCustom (reduction = "tsne" ,pt.size = 0.5 ,group.by = "ClusterName" ,label = TRUE ,repel = TRUE ,figure_plot = TRUE ,color_seed = reseed& NoLegend ()
UMAP
Code
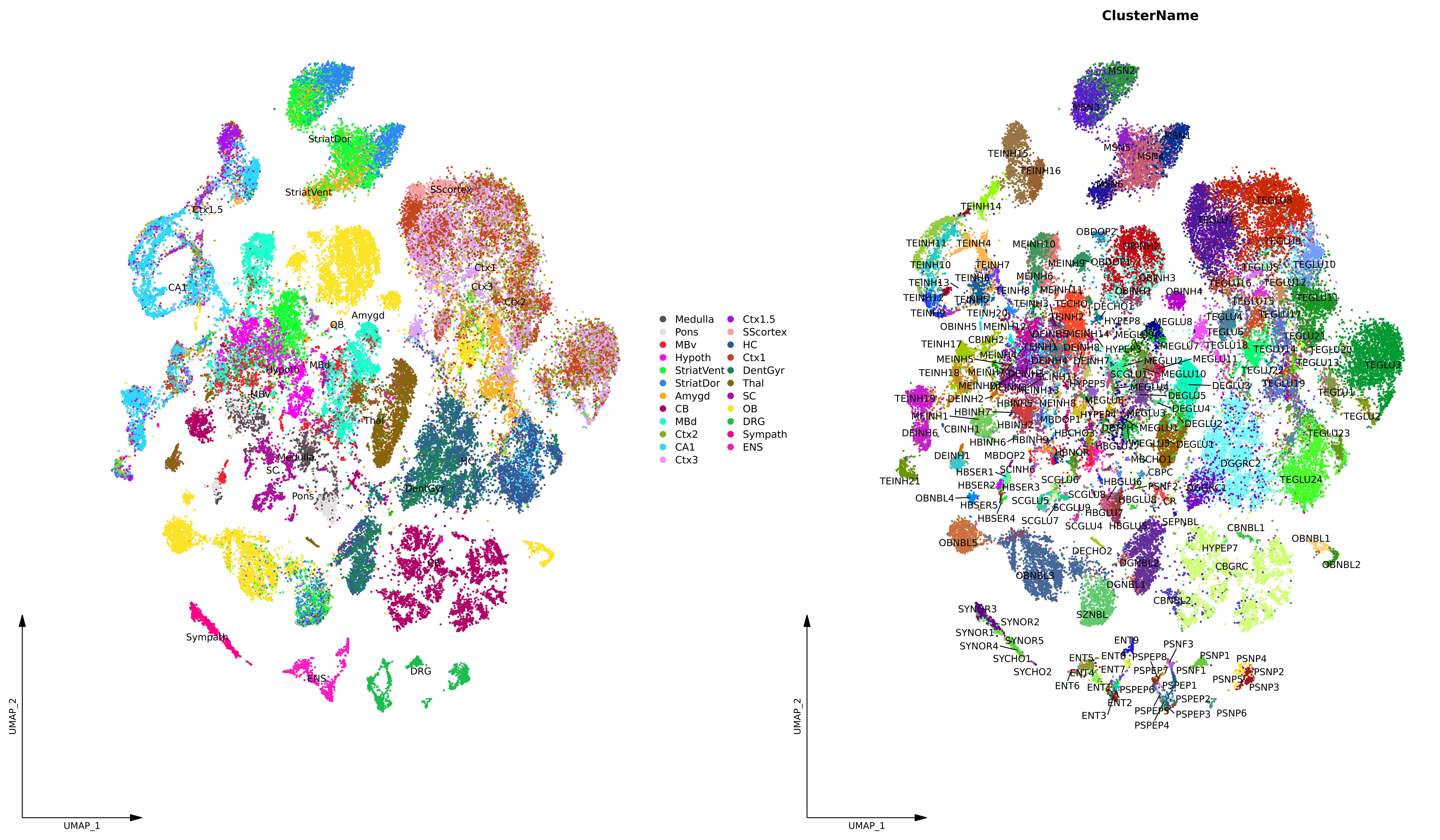
DimPlot_scCustom (reduction = "umap" ,pt.size = 0.5 ,label = TRUE ,repel = TRUE ,figure_plot = TRUE ,color_seed = reseed| DimPlot_scCustom (reduction = "umap" ,pt.size = 0.5 ,group.by = "ClusterName" ,label = TRUE ,repel = TRUE ,figure_plot = TRUE ,color_seed = reseed& NoLegend ()
Code
FeaturePlot_scCustom (l6.srt, goi, num_columns = 1 , pt.size = 1 , order = T, colors_use = viridis:: viridis (n = 100 , alpha = .6 , direction = - 1 , option = "E" ))
Code
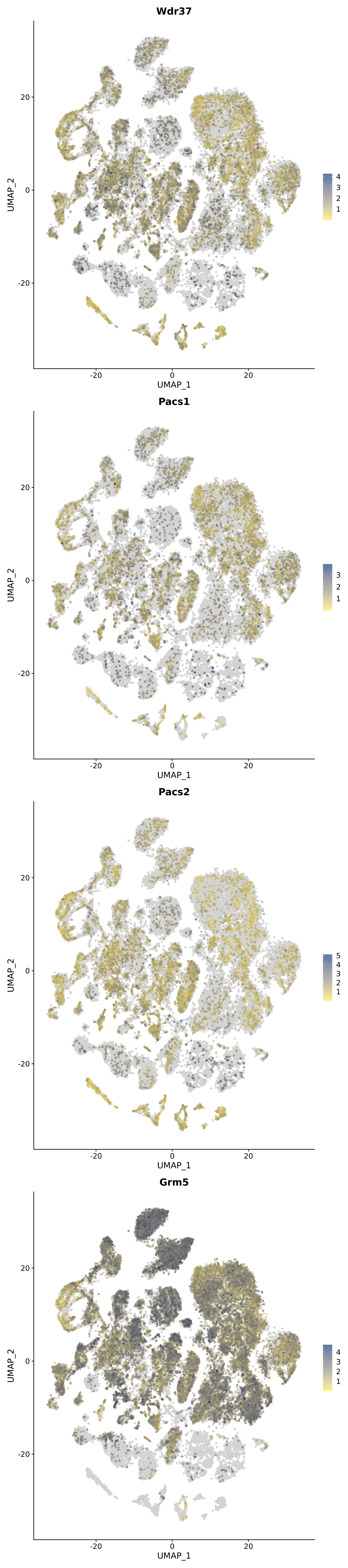
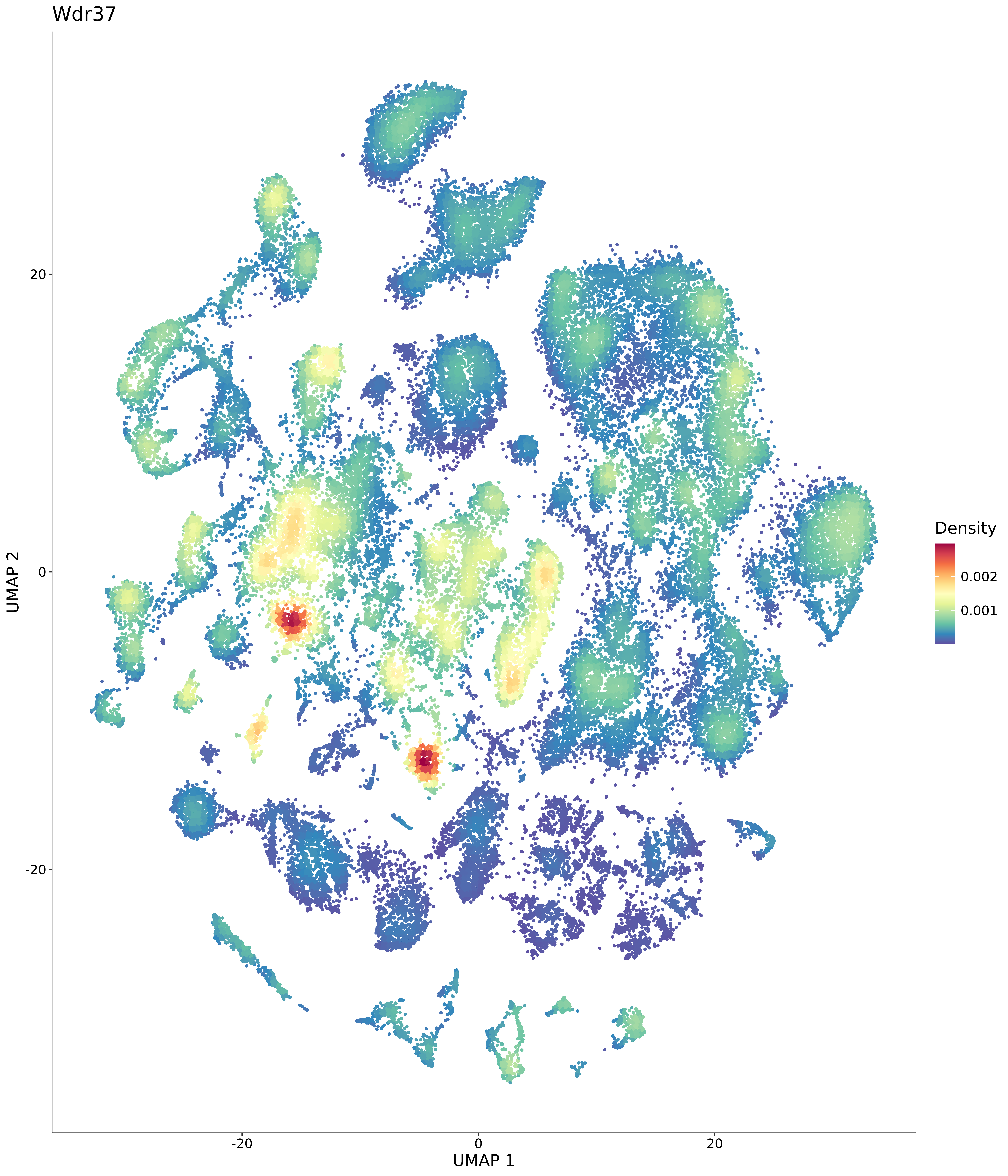
Plot_Density_Custom (seurat_object = l6.srt, features = "Wdr37" , custom_palette = l6.srt@ misc$ div_Colour_Pal)
Code
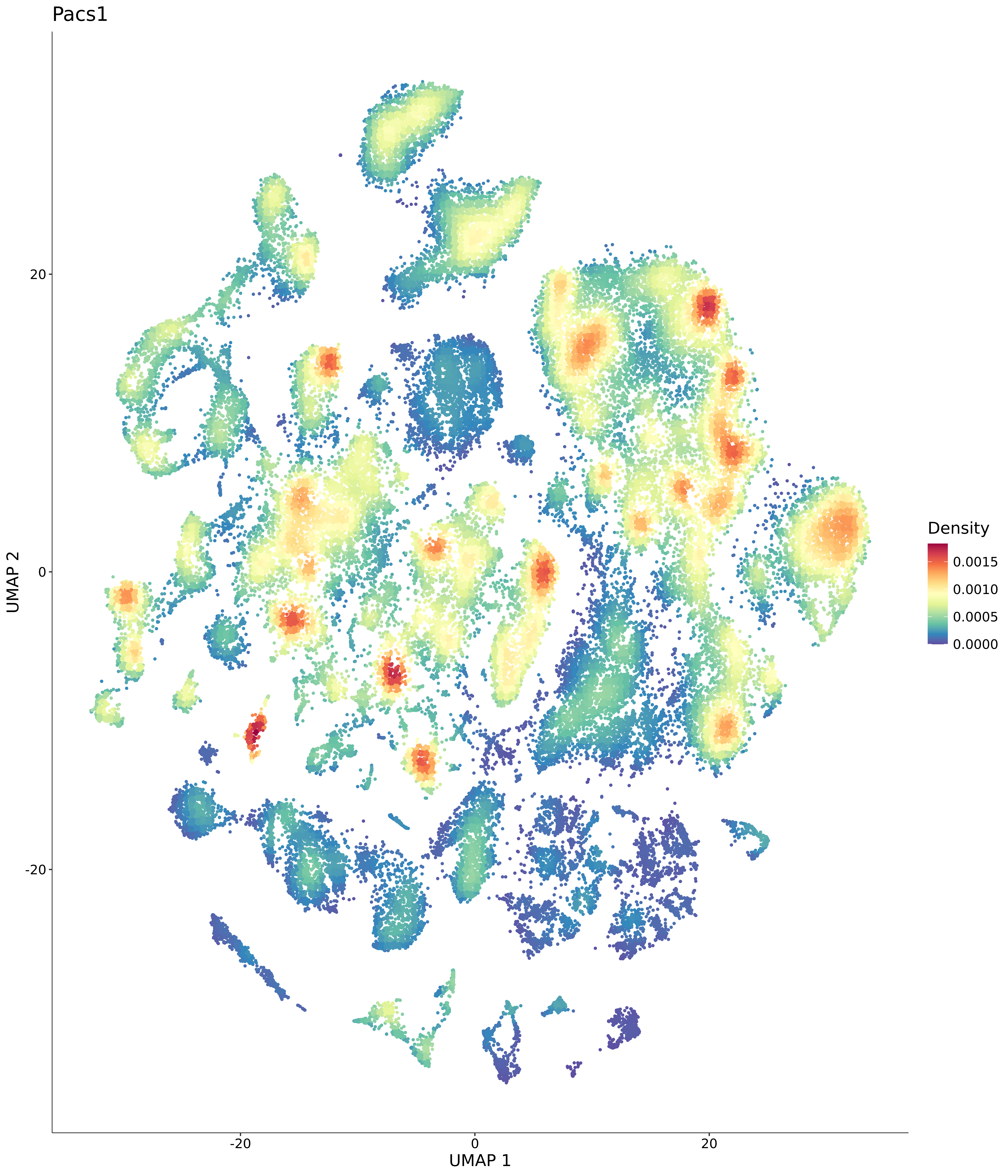
Plot_Density_Custom (seurat_object = l6.srt, features = "Pacs1" , custom_palette = l6.srt@ misc$ div_Colour_Pal)
Code
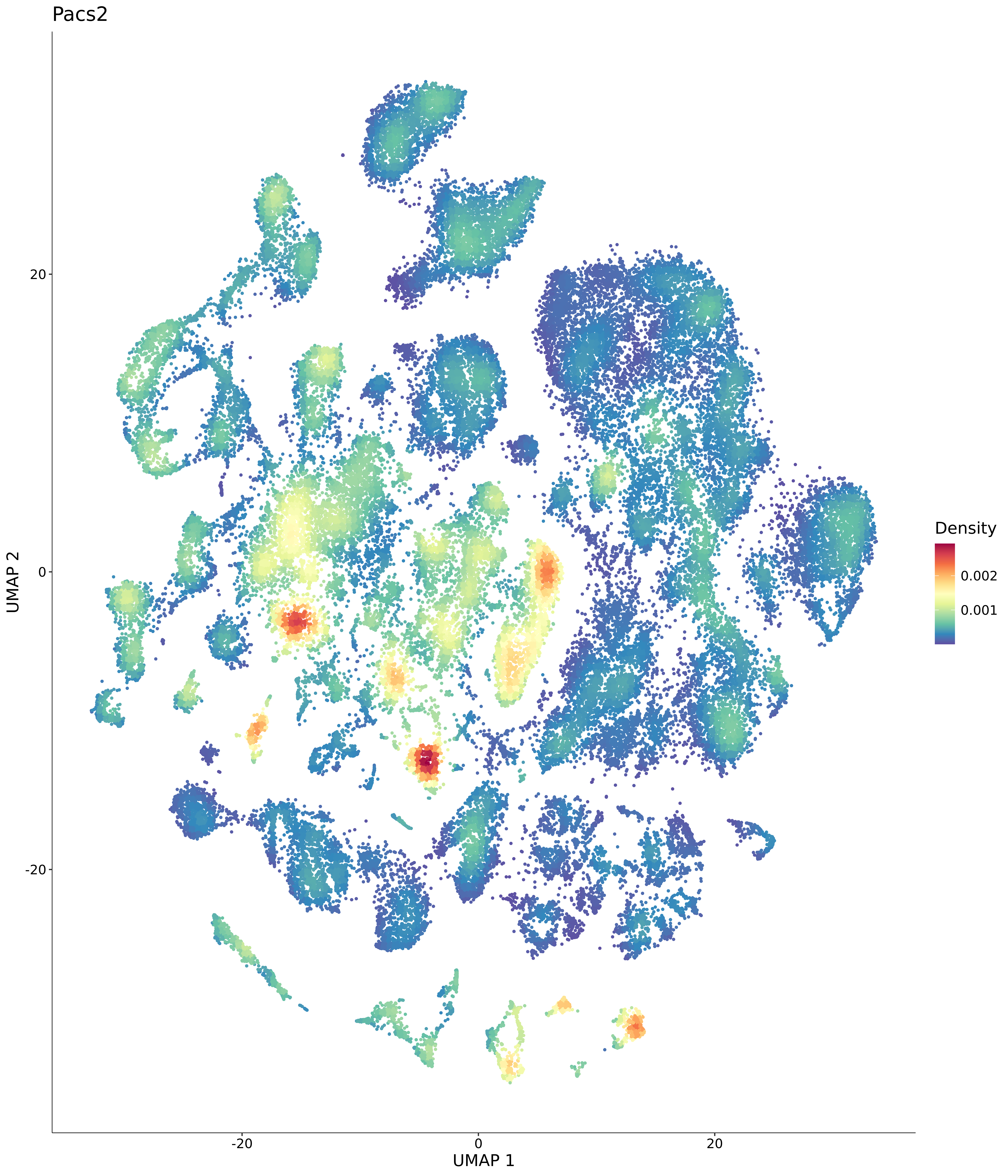
Plot_Density_Custom (seurat_object = l6.srt, features = "Pacs2" , custom_palette = l6.srt@ misc$ div_Colour_Pal)
Code
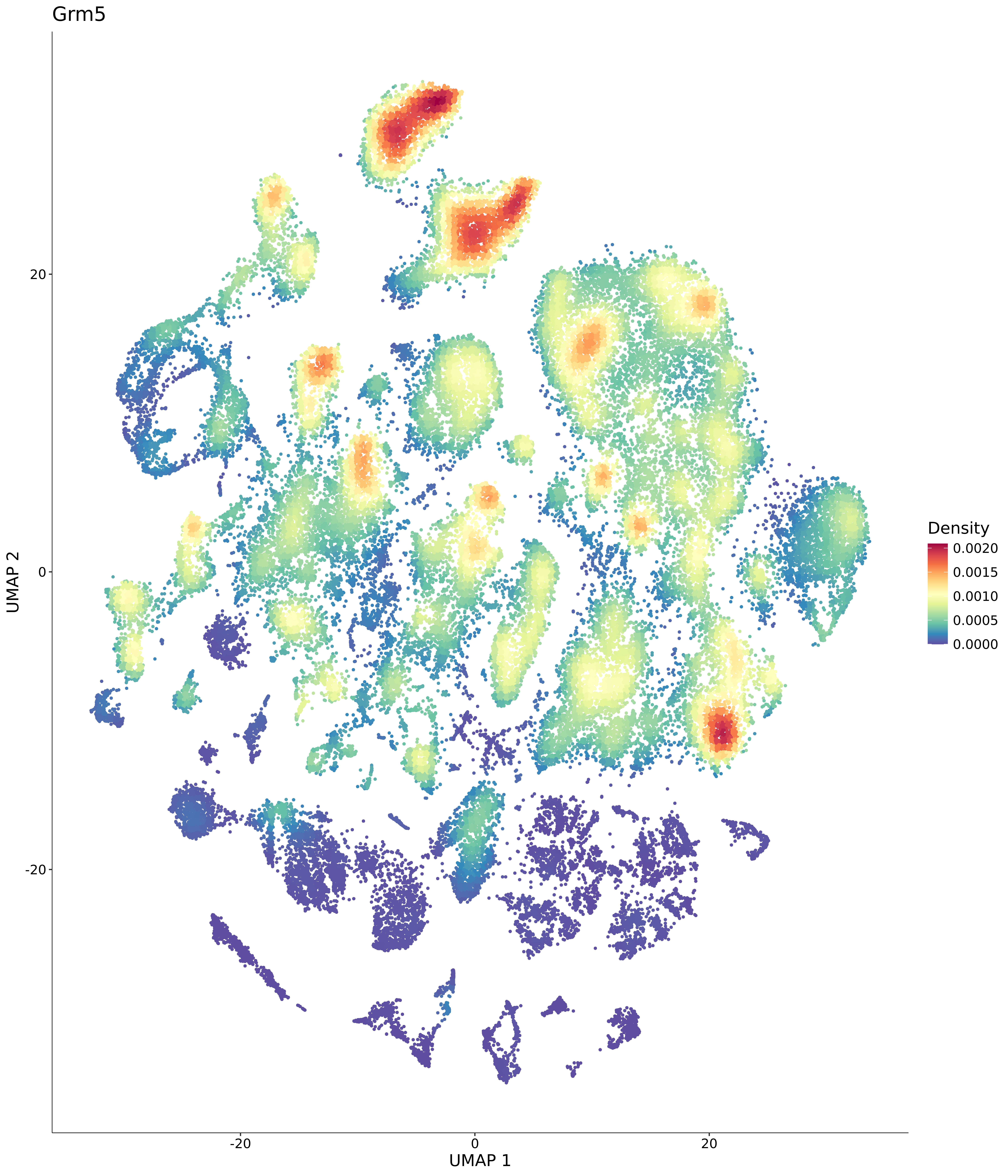
Plot_Density_Custom (seurat_object = l6.srt, features = "Grm5" , custom_palette = l6.srt@ misc$ div_Colour_Pal)
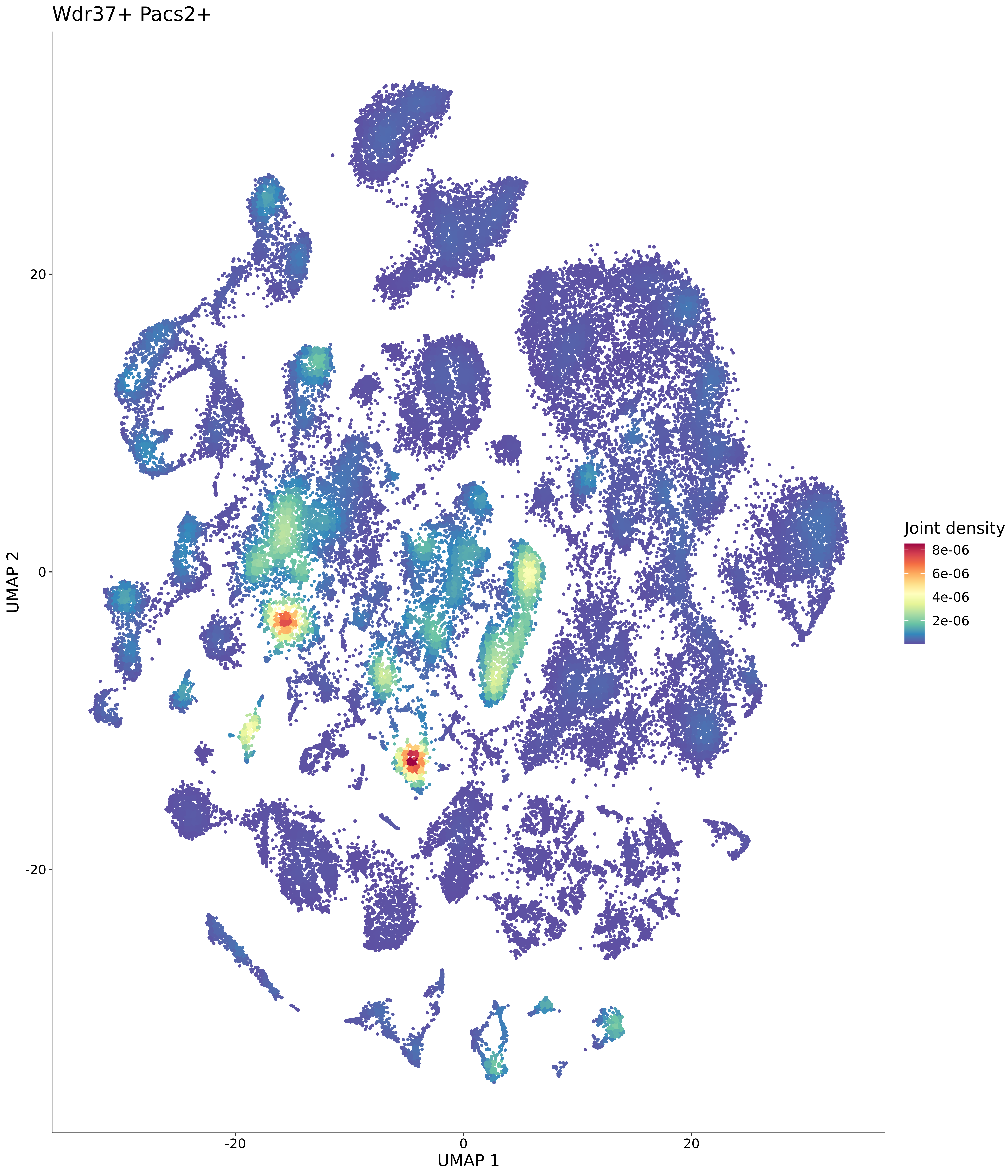
UMAP density plots of interactions between Wdr37 and other features
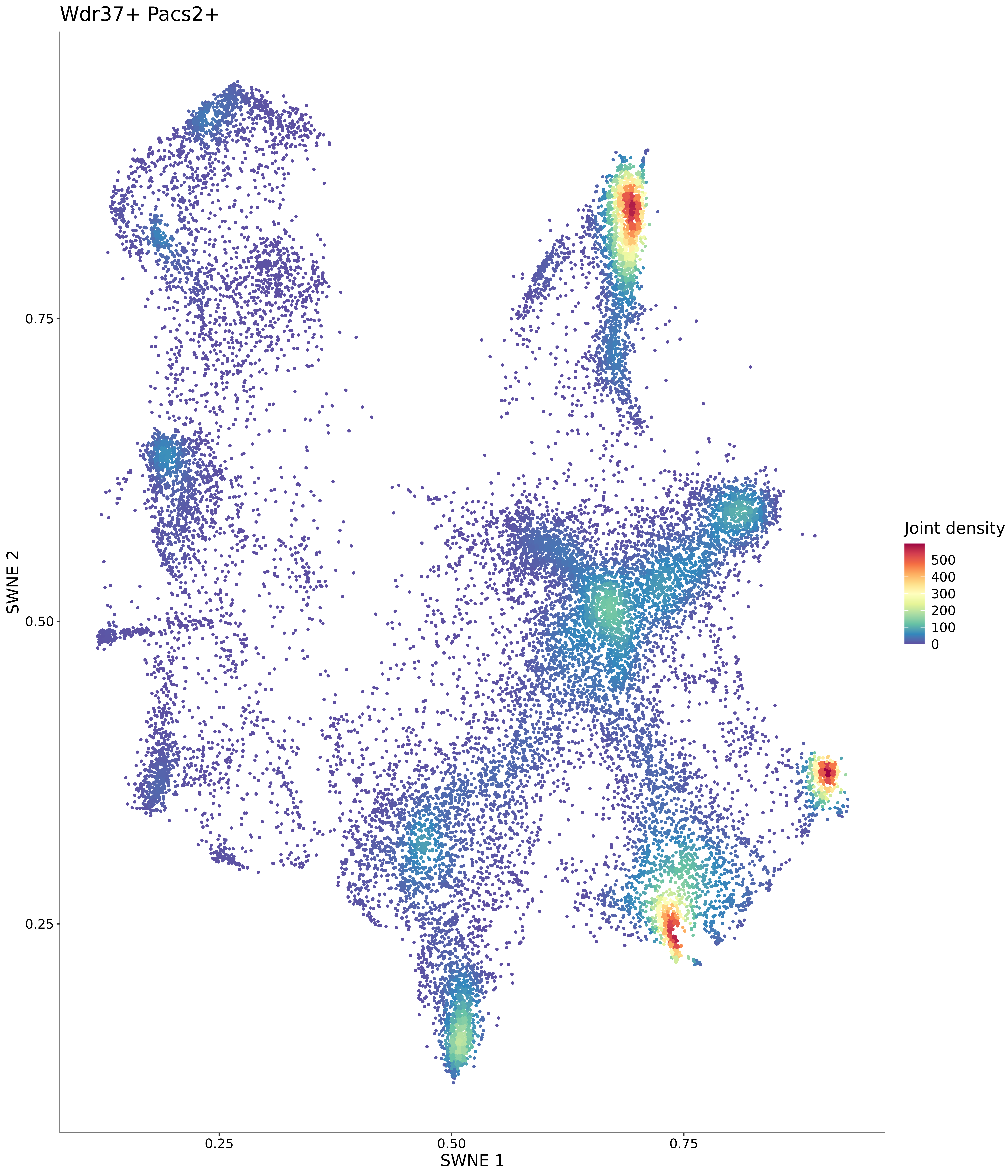
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Wdr37" , "Pacs2" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
Code
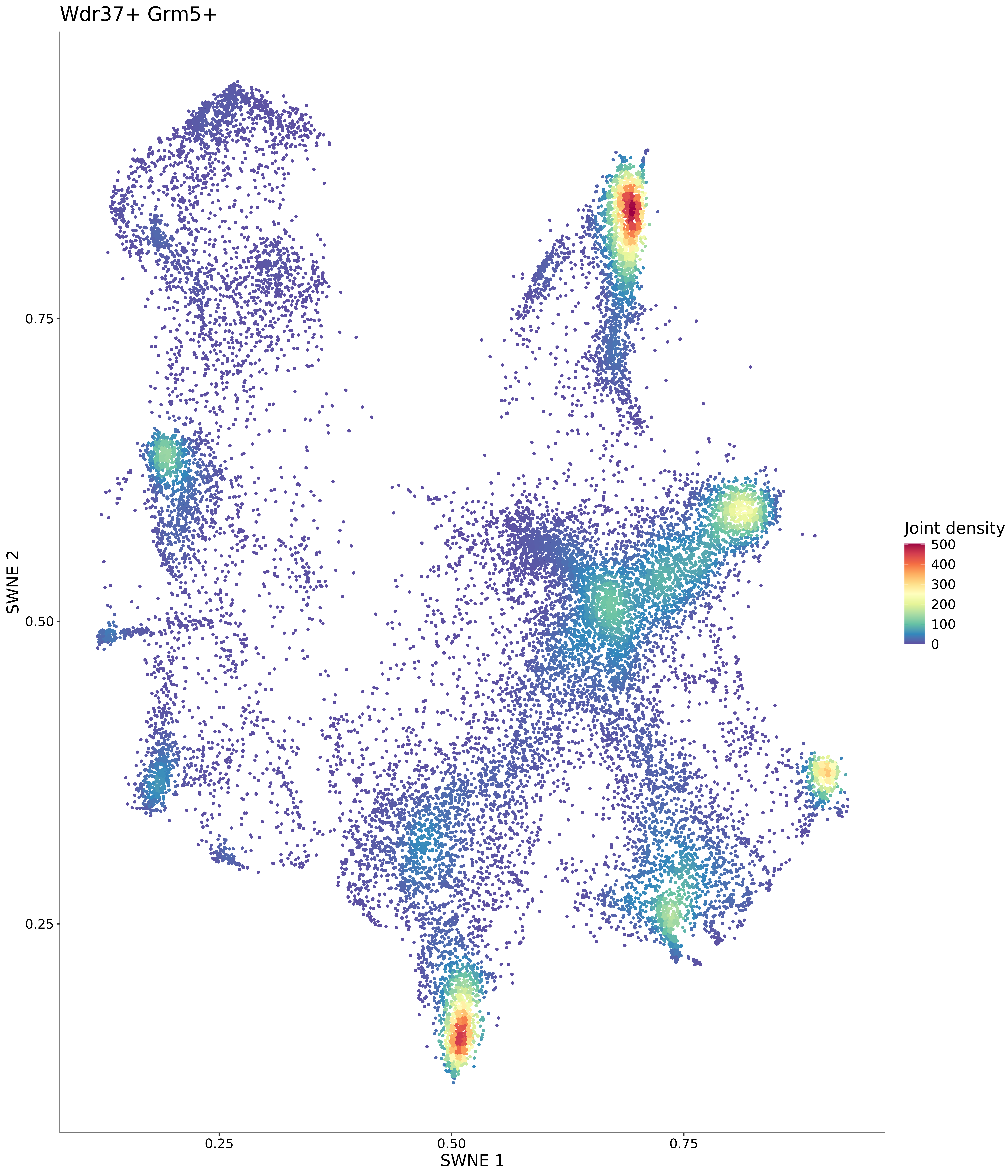
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Wdr37" , "Grm5" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
Code
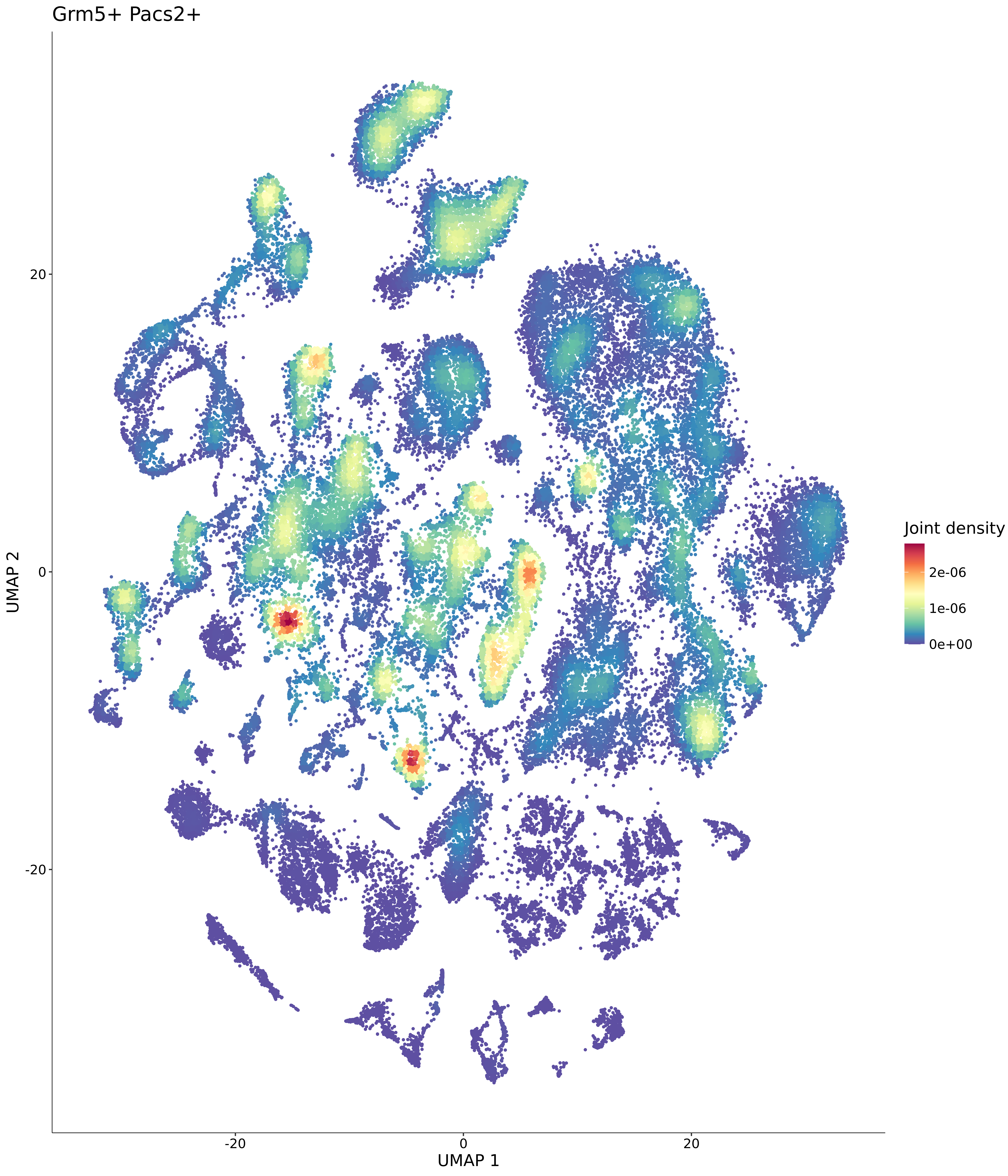
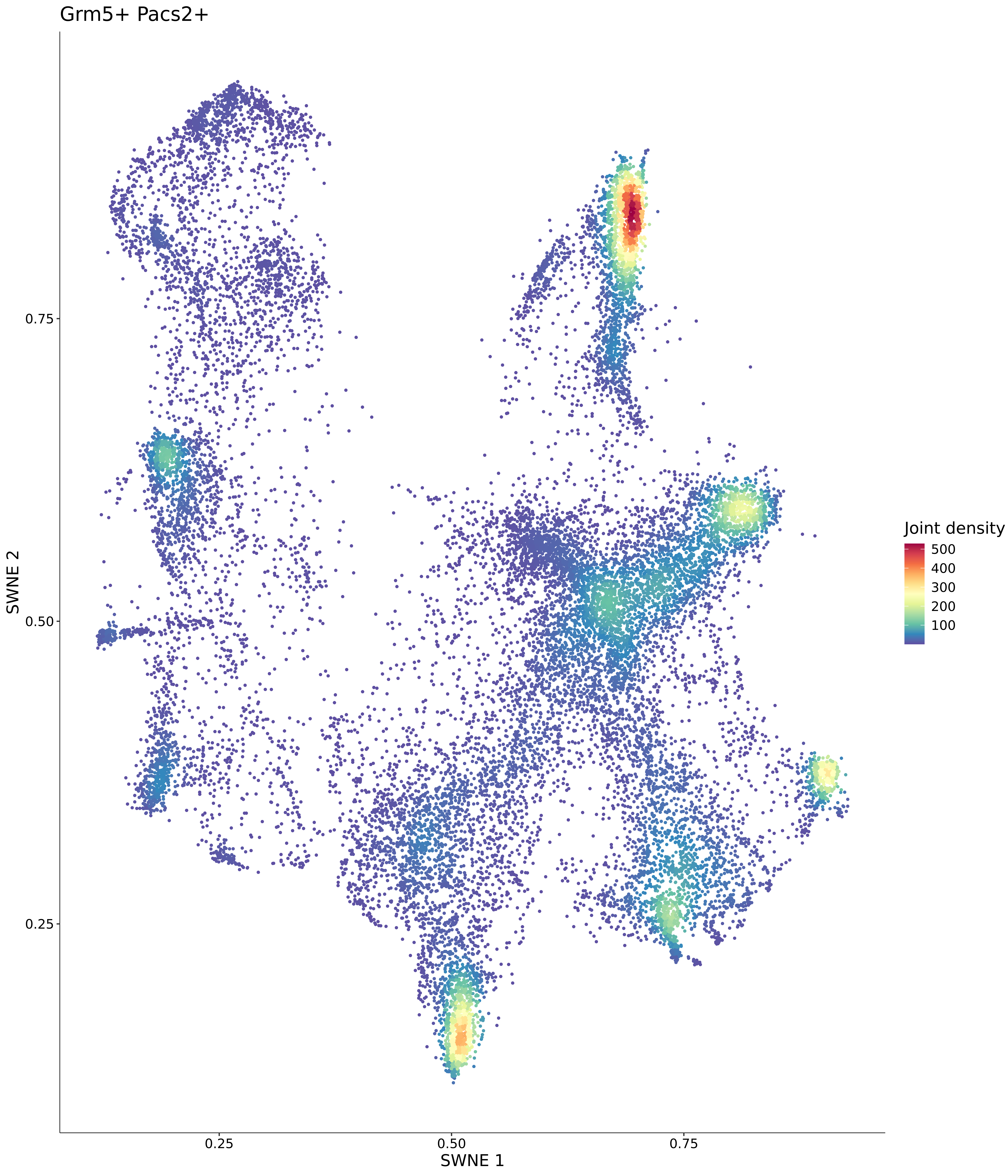
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Grm5" , "Pacs2" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
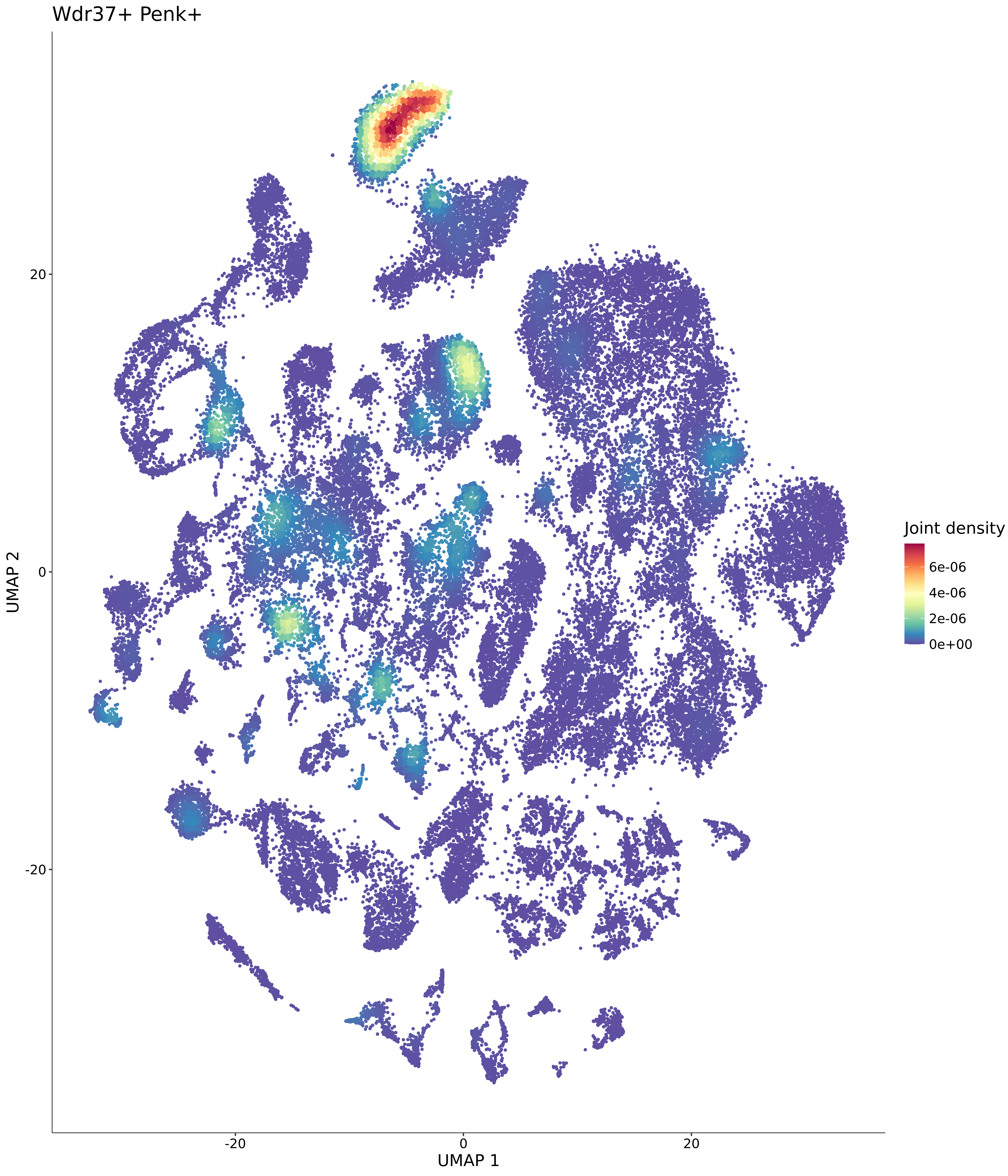
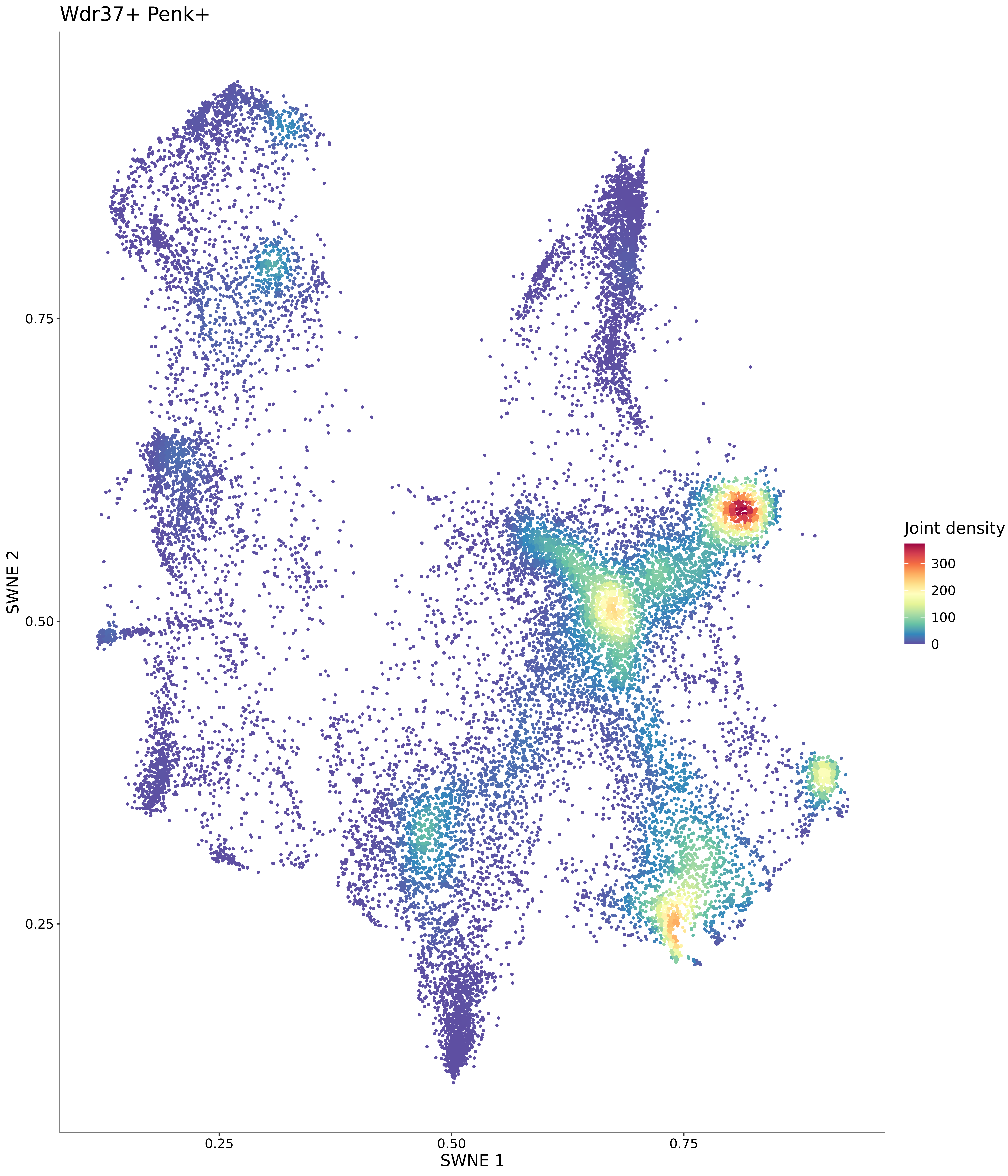
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Wdr37" , "Penk" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
Code
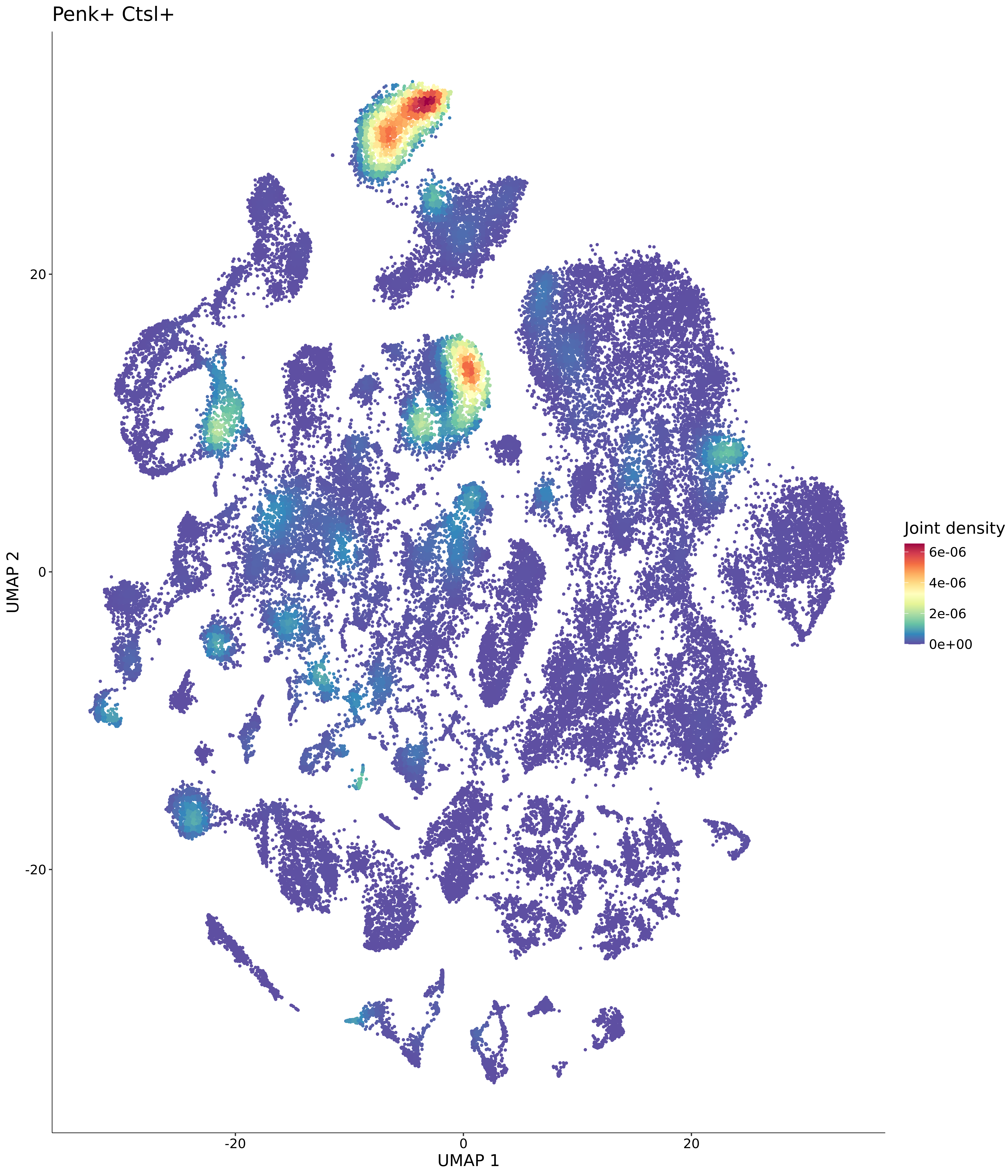
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Penk" , "Ctsl" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
UMAP density plots of neurotransmitter phenotype with Wdr37 and other features
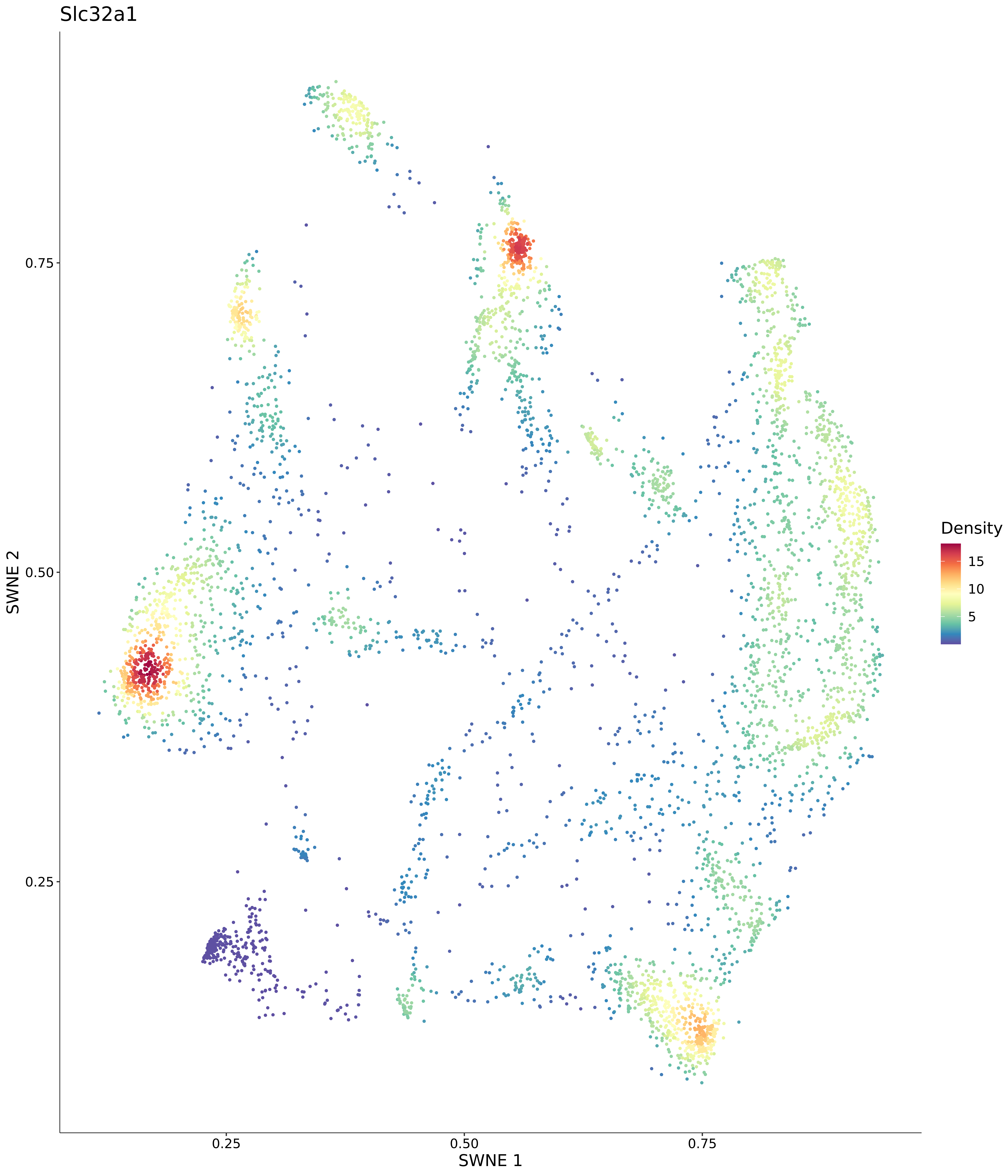
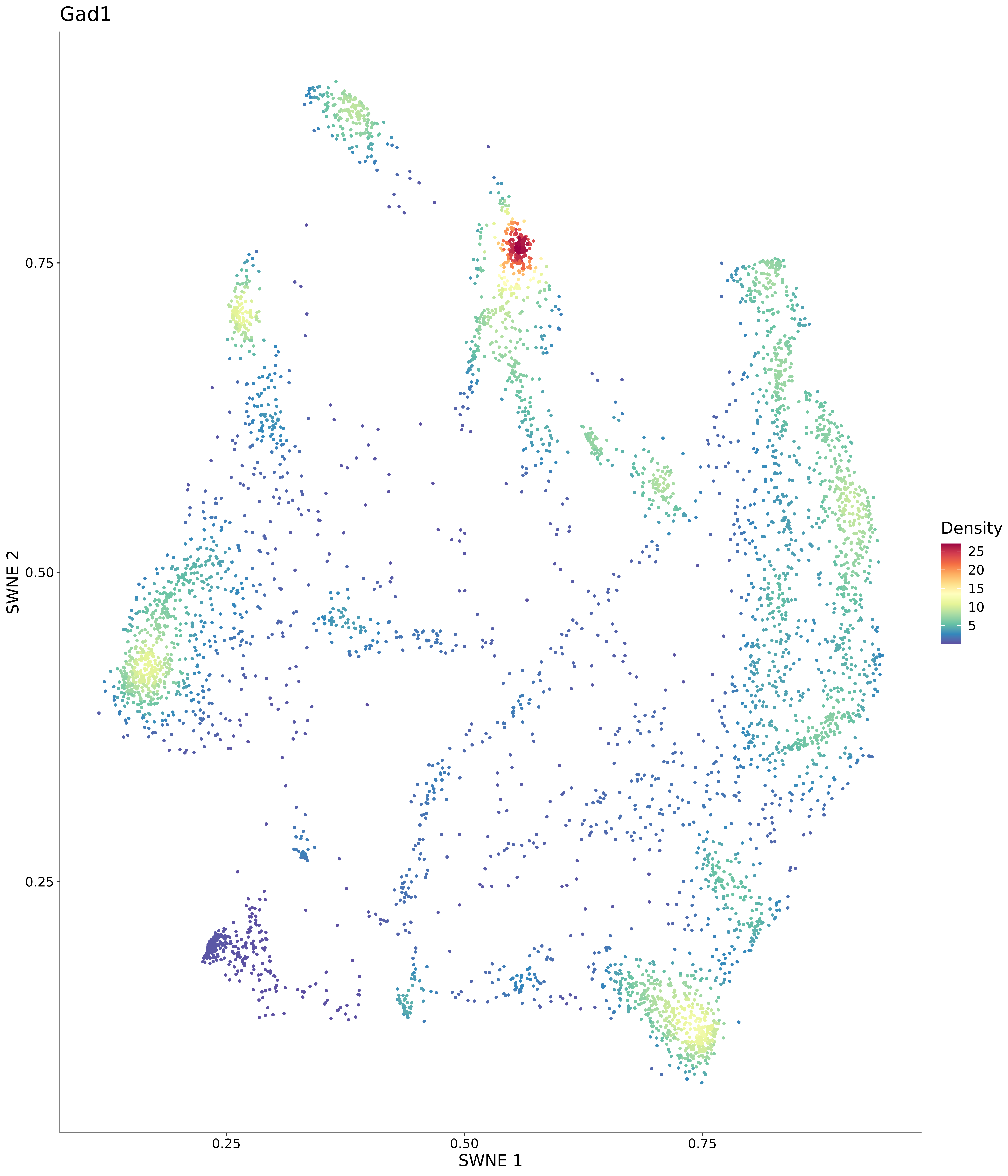
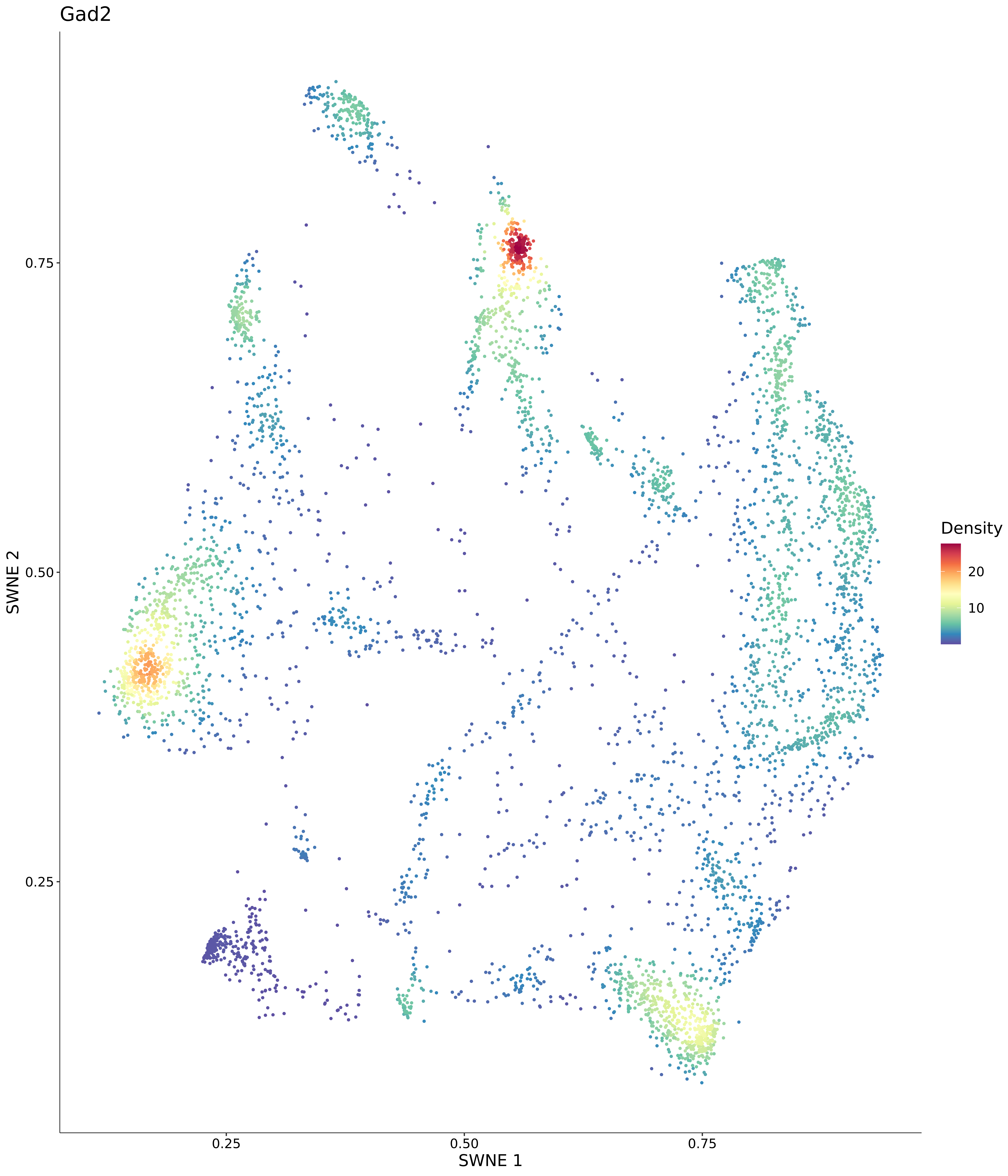
GABA
Code
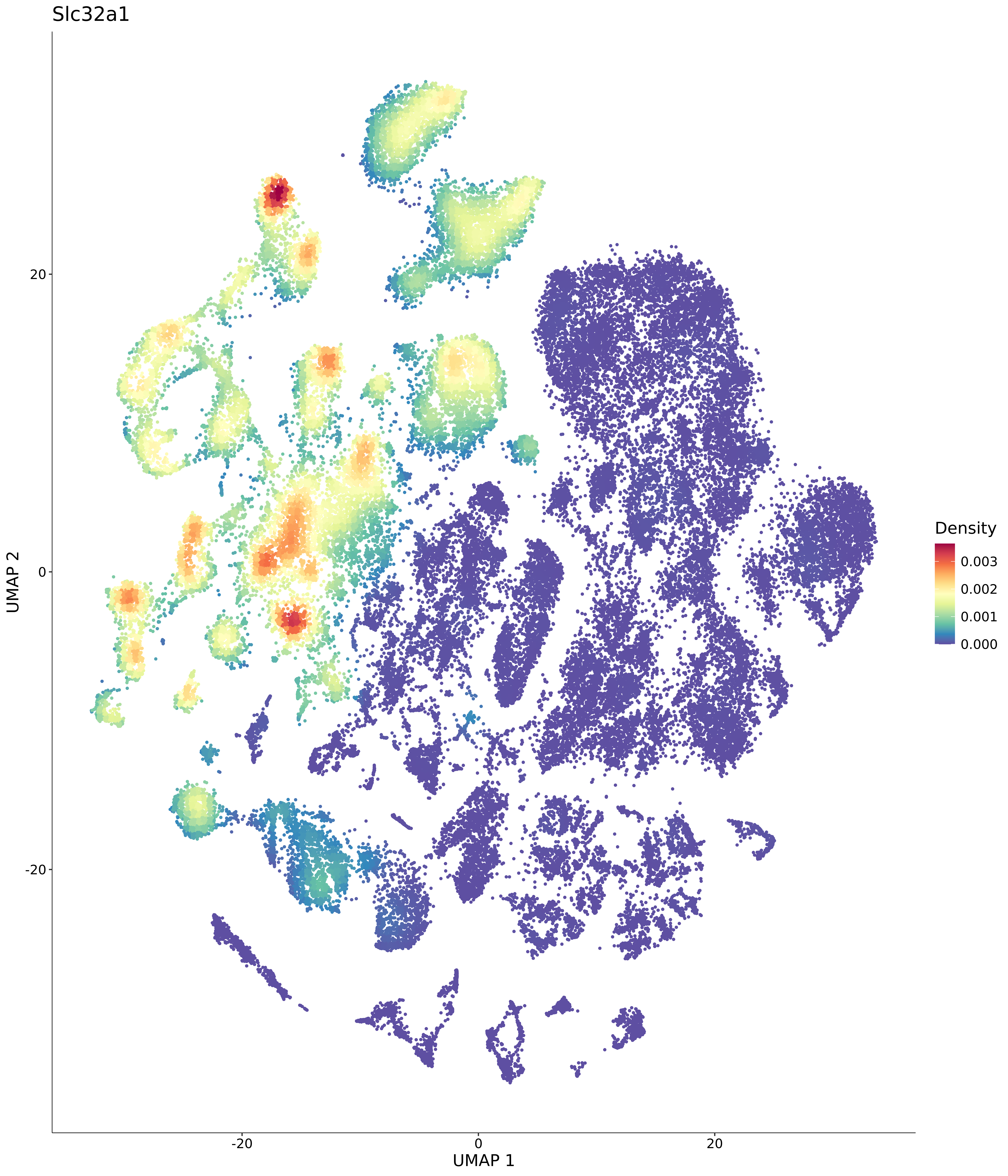
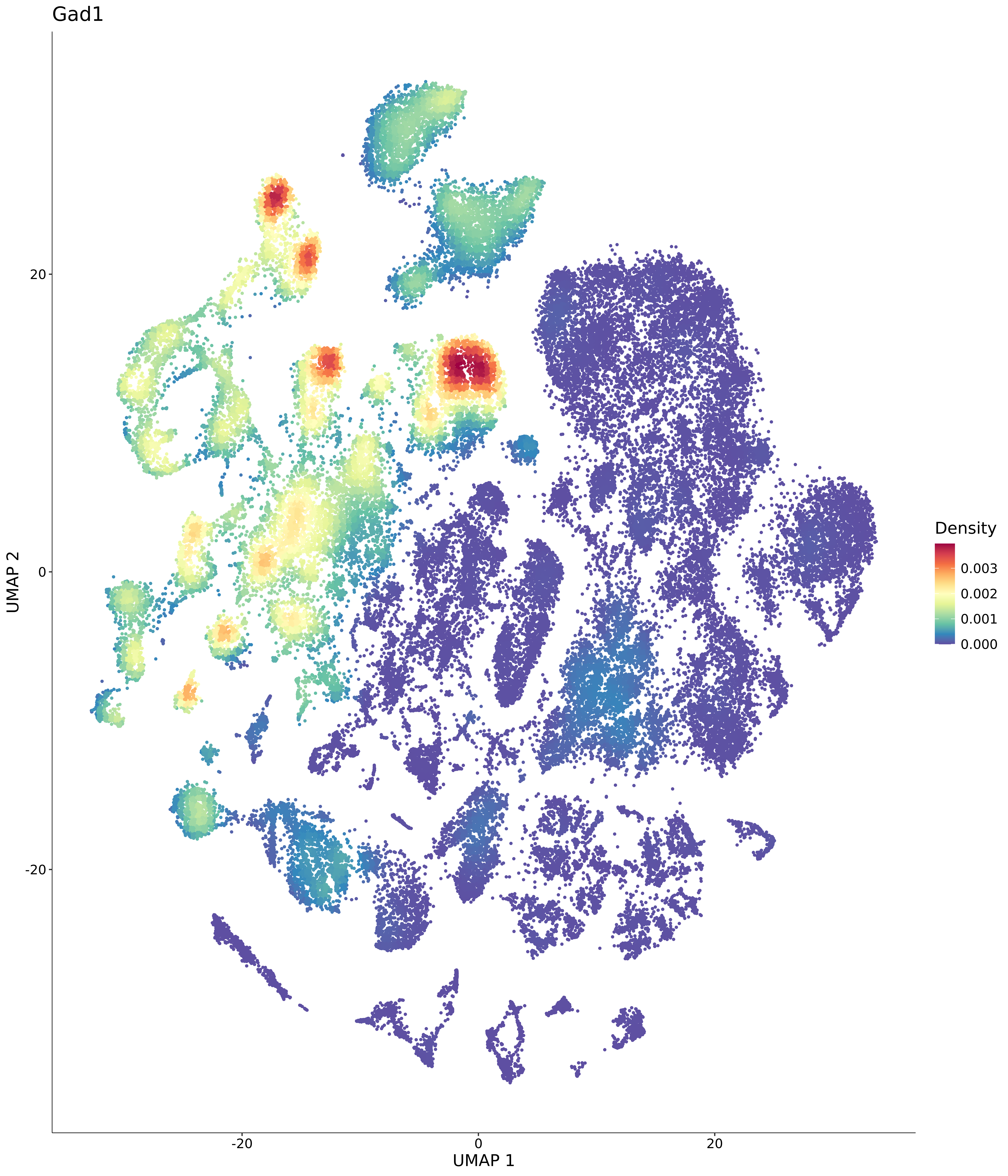
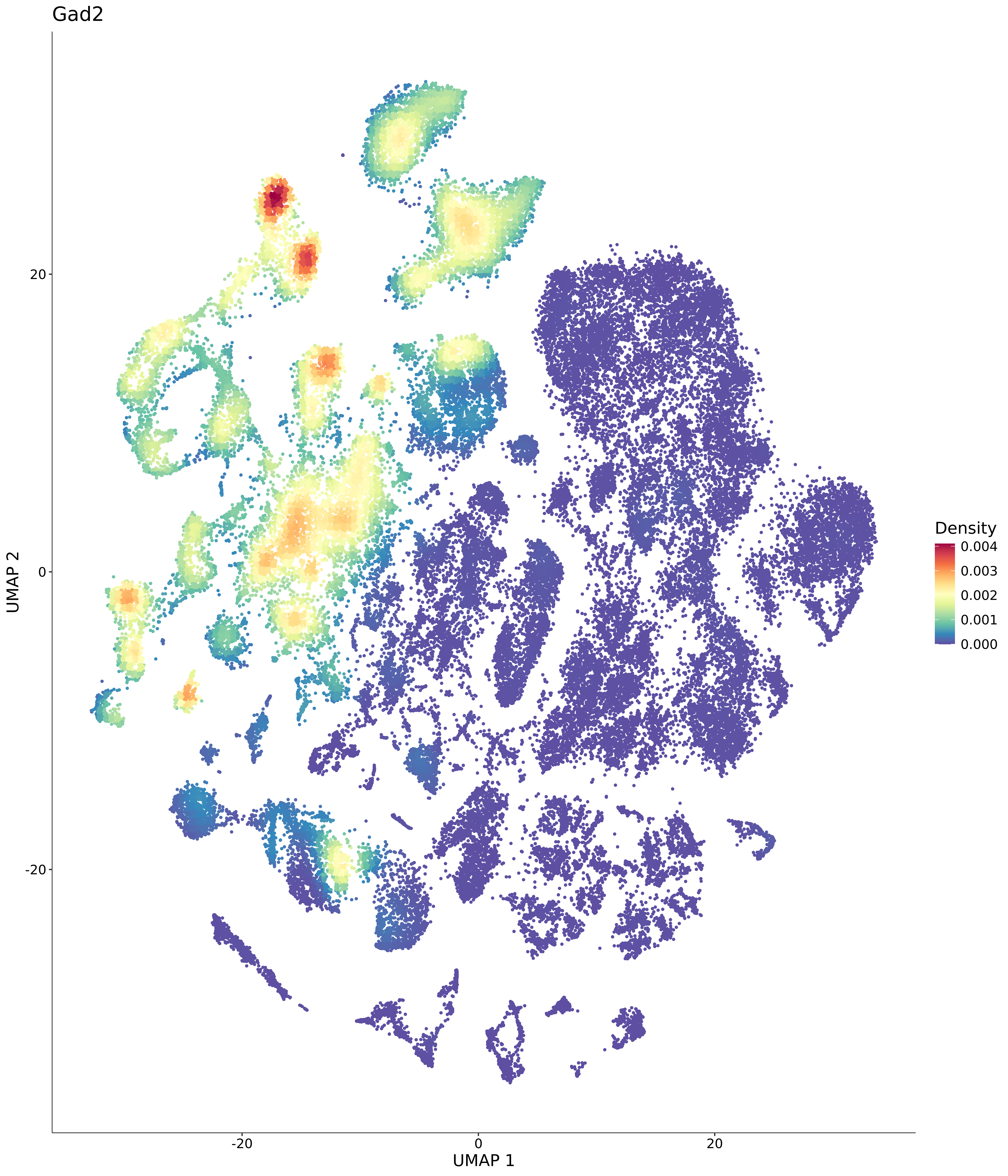
Plot_Density_Custom (seurat_object = l6.srt, features = c ("Slc32a1" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
Code
Plot_Density_Custom (seurat_object = l6.srt, features = c ("Gad1" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
Code
Plot_Density_Custom (seurat_object = l6.srt, features = c ("Gad2" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
Glutamate
Code
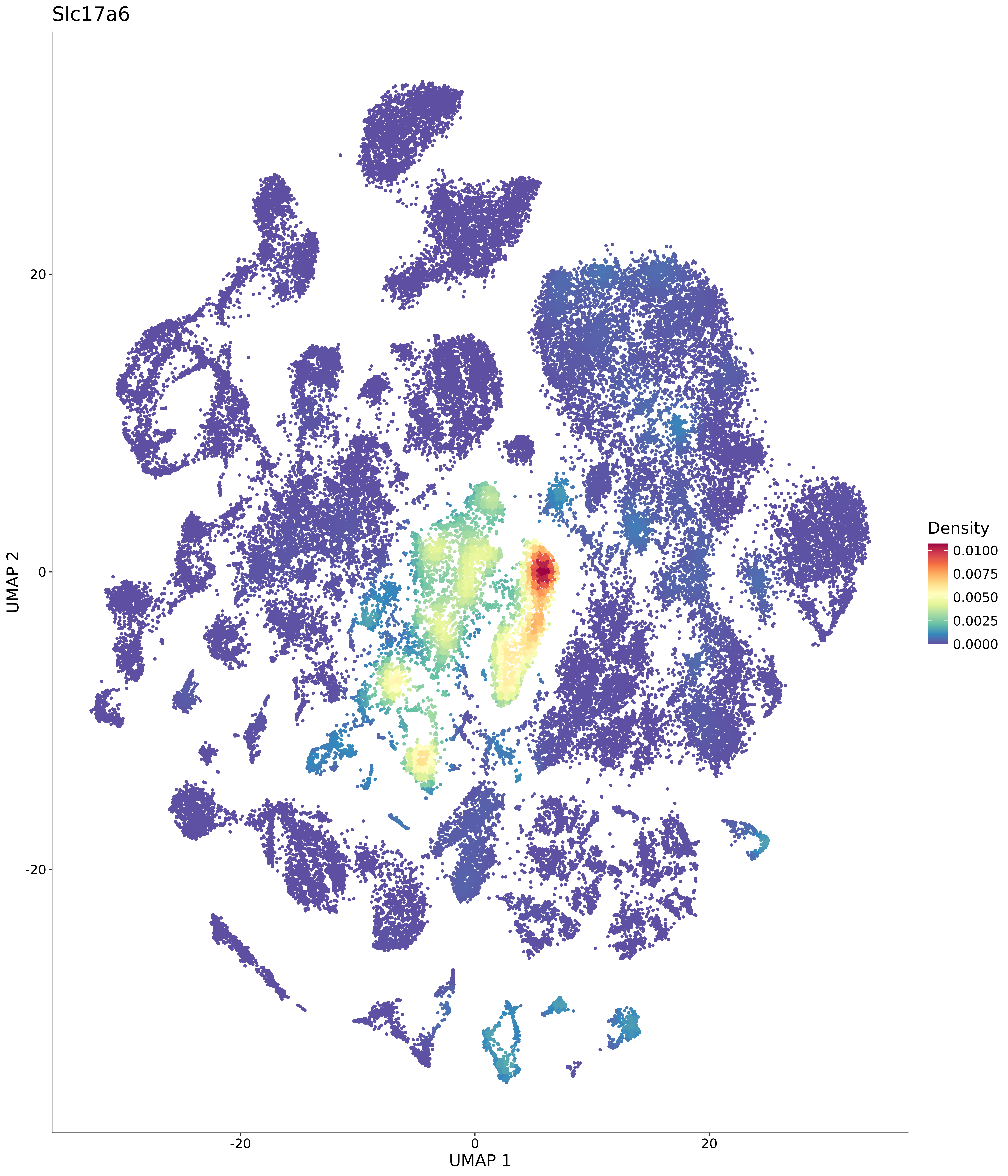
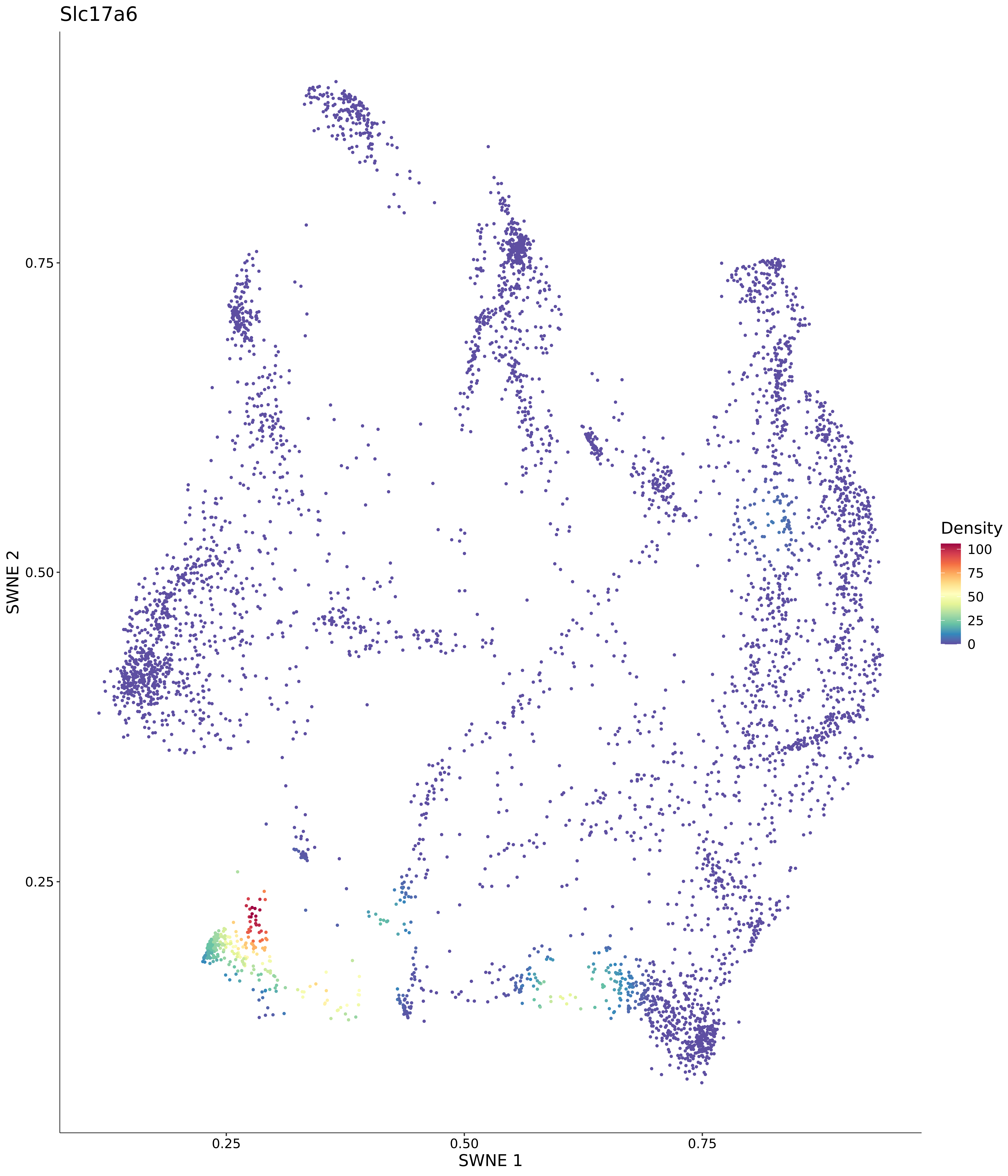
Plot_Density_Custom (seurat_object = l6.srt, features = c ("Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
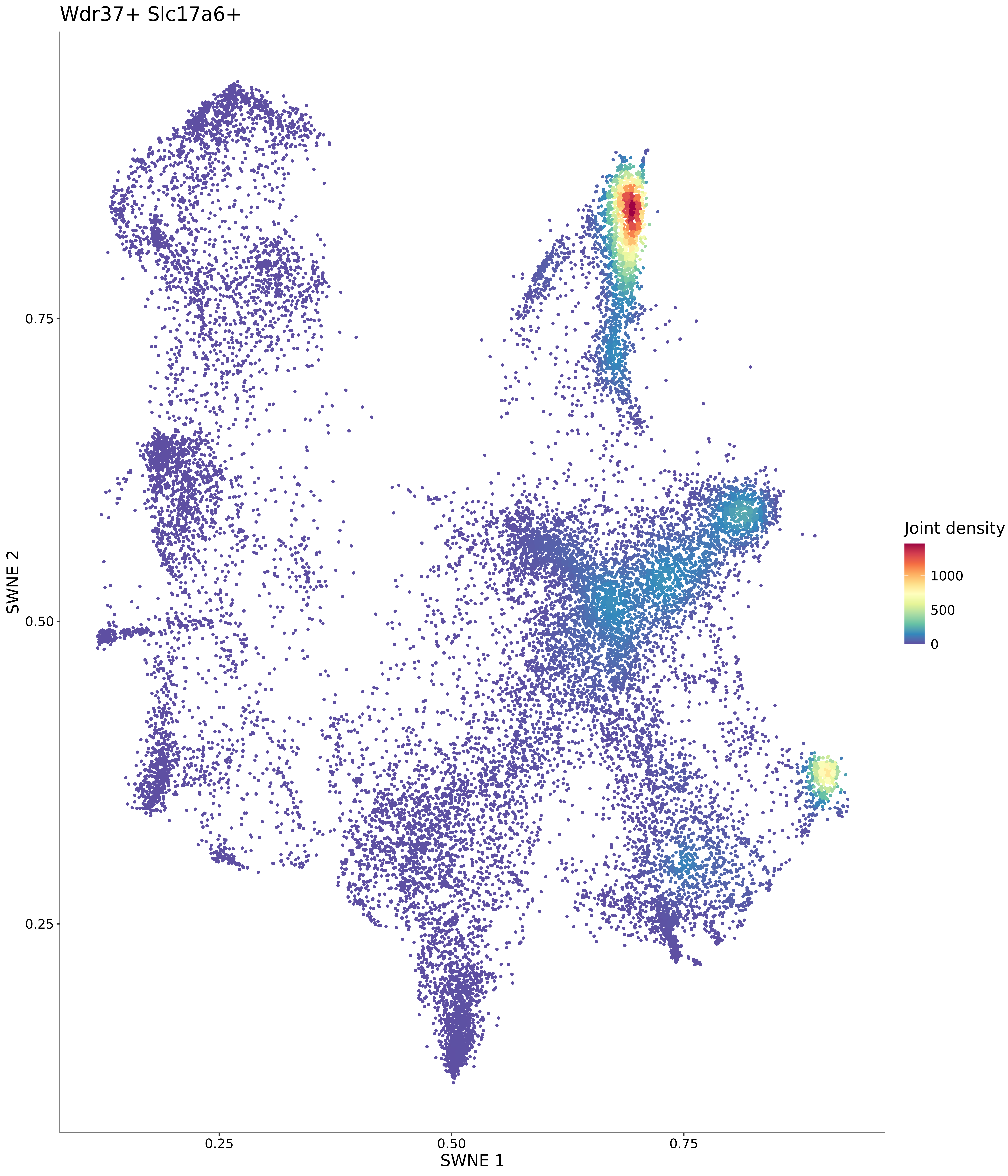
Code
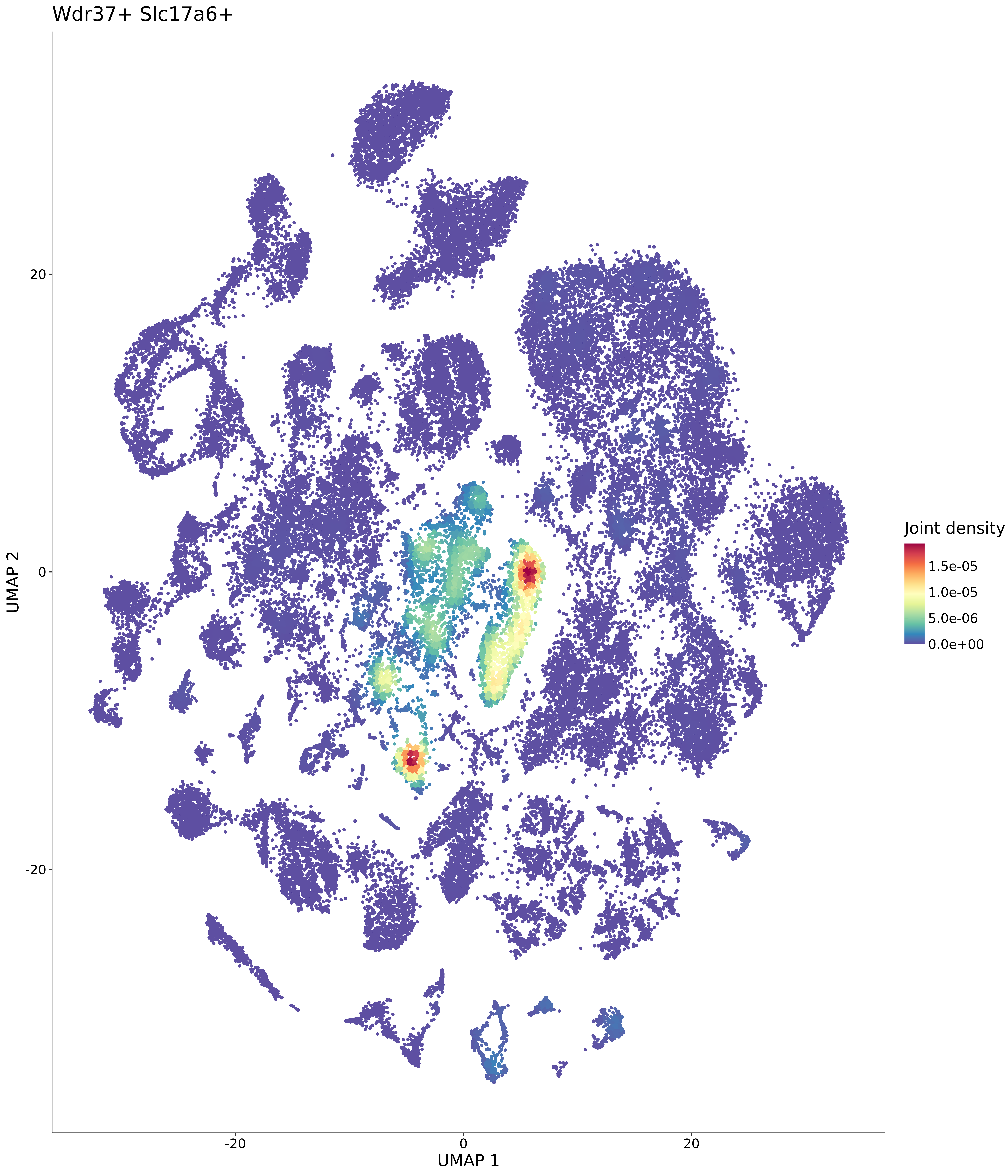
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Wdr37" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
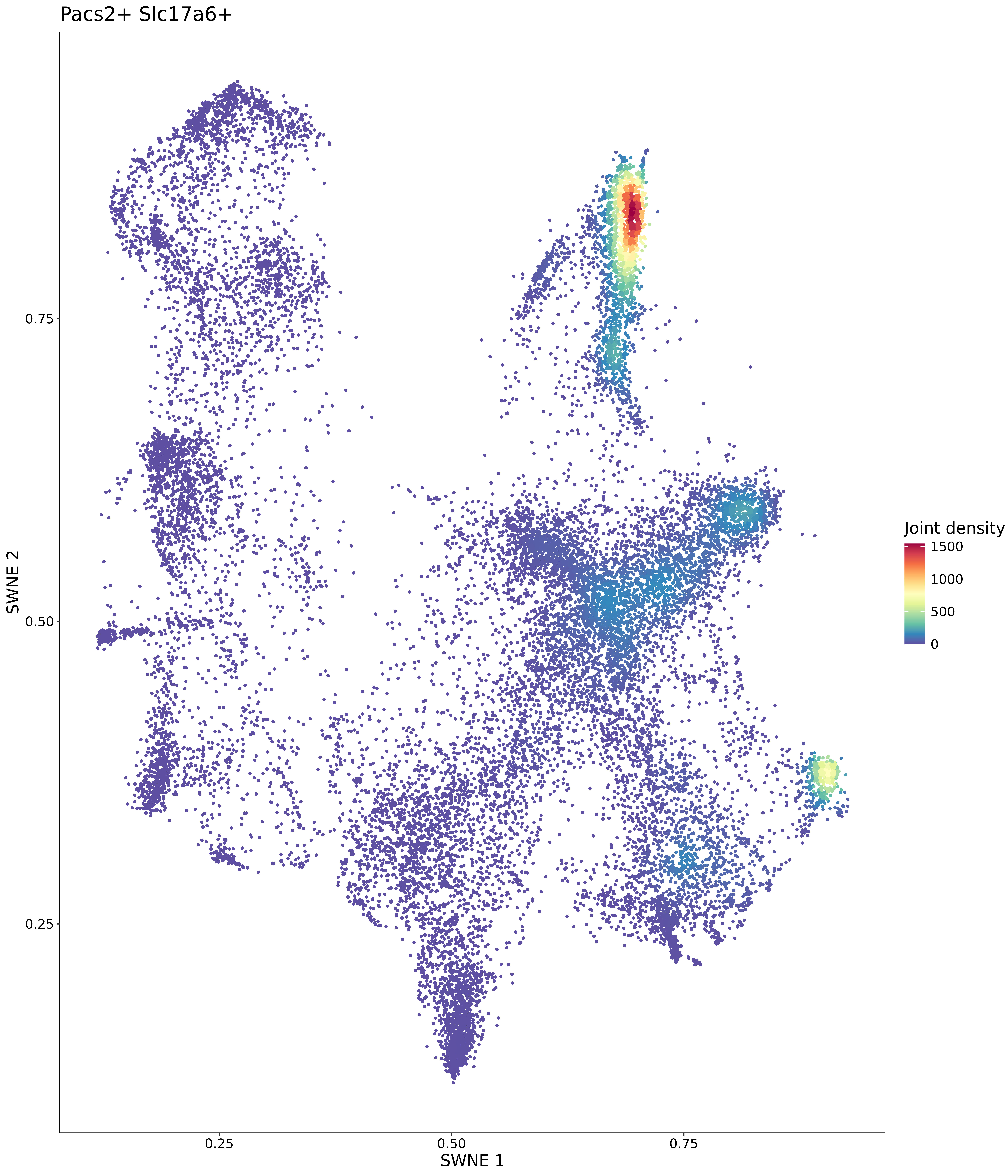
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Pacs2" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
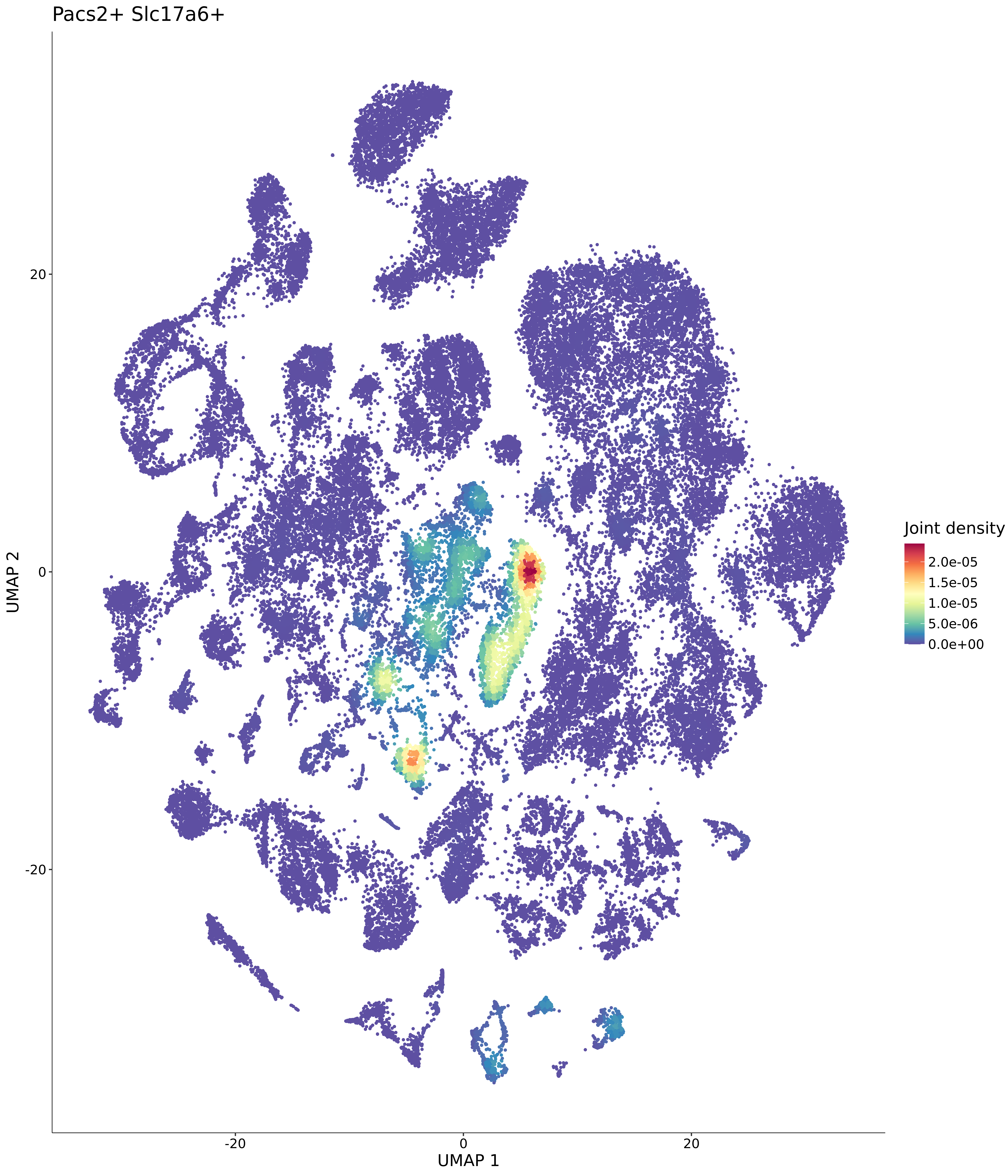
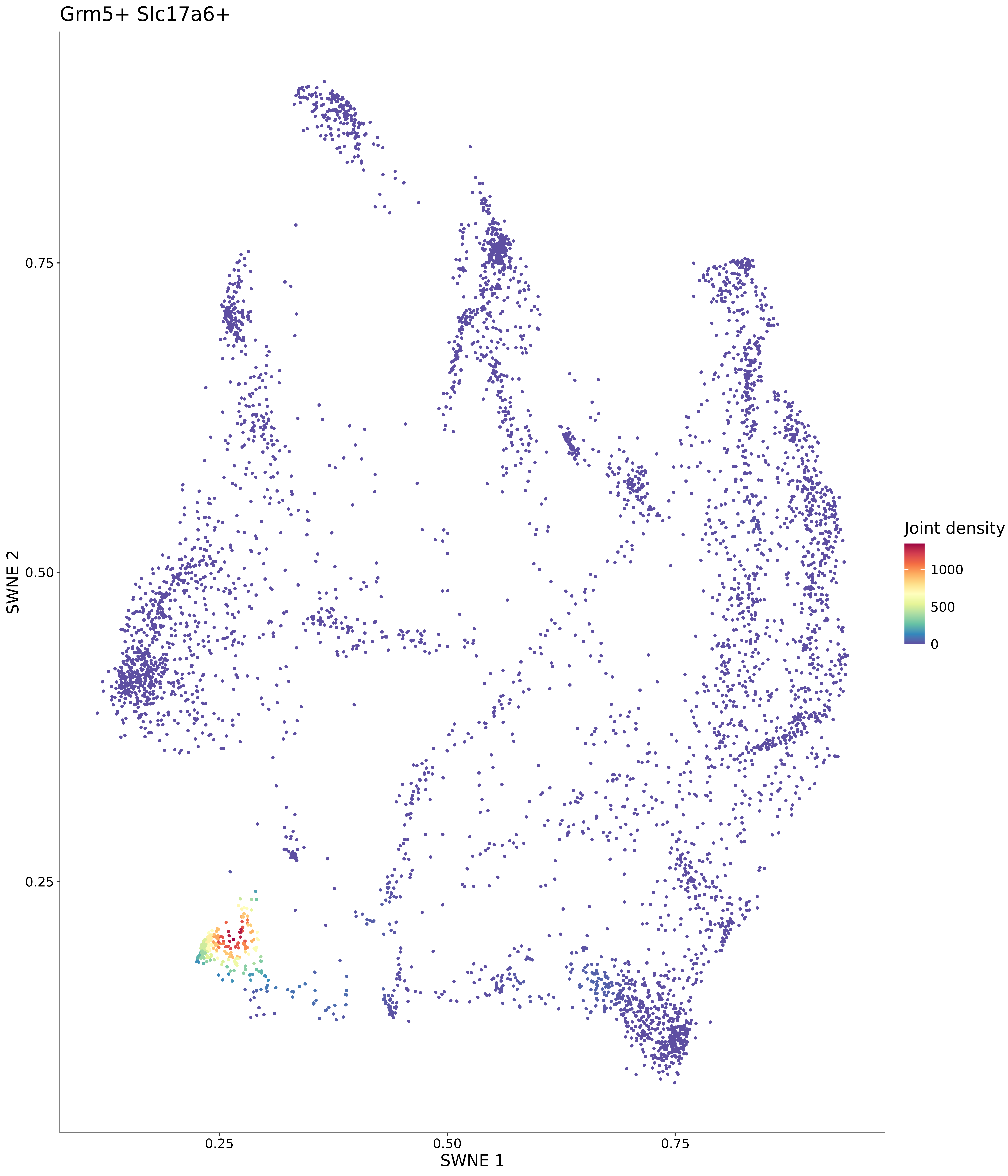
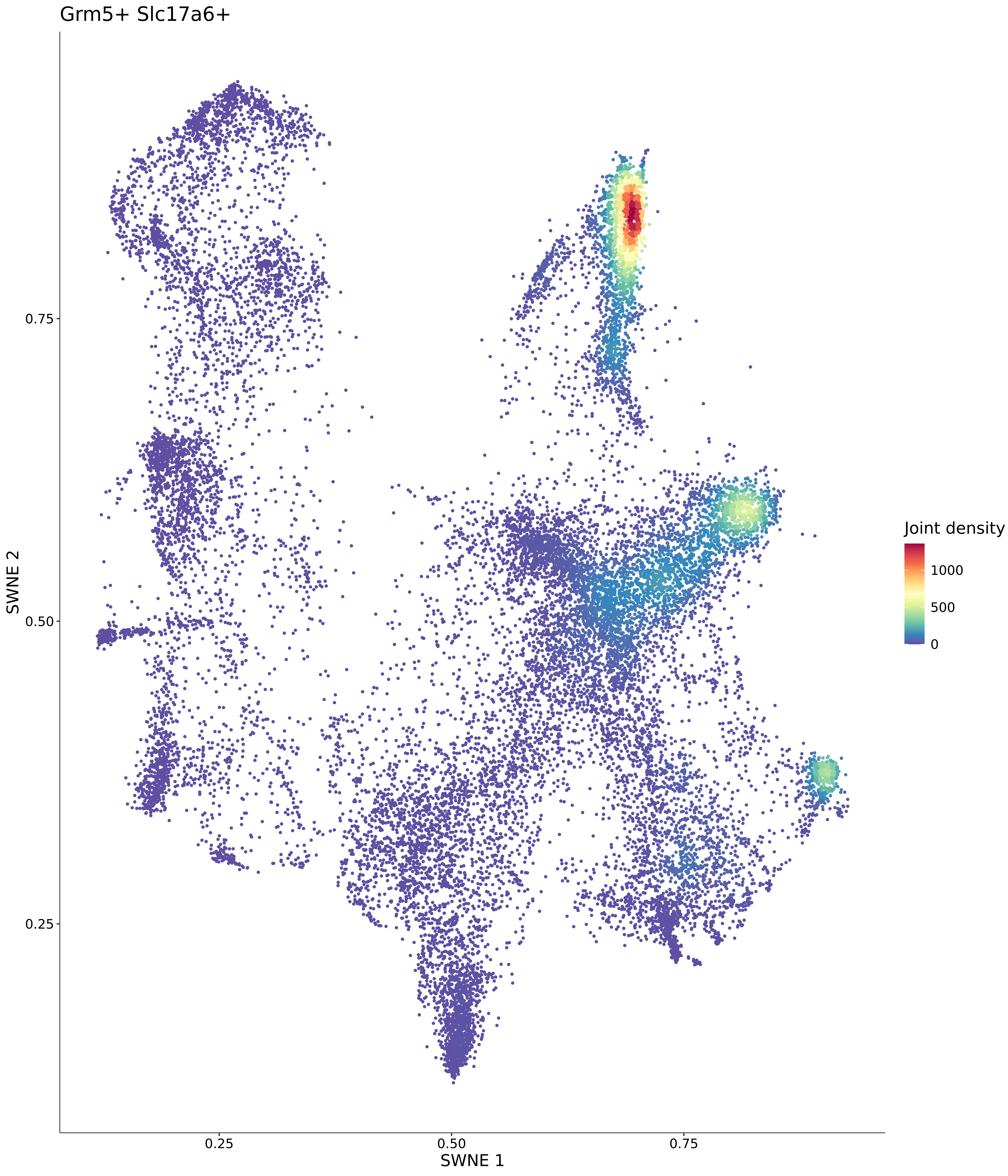
Code
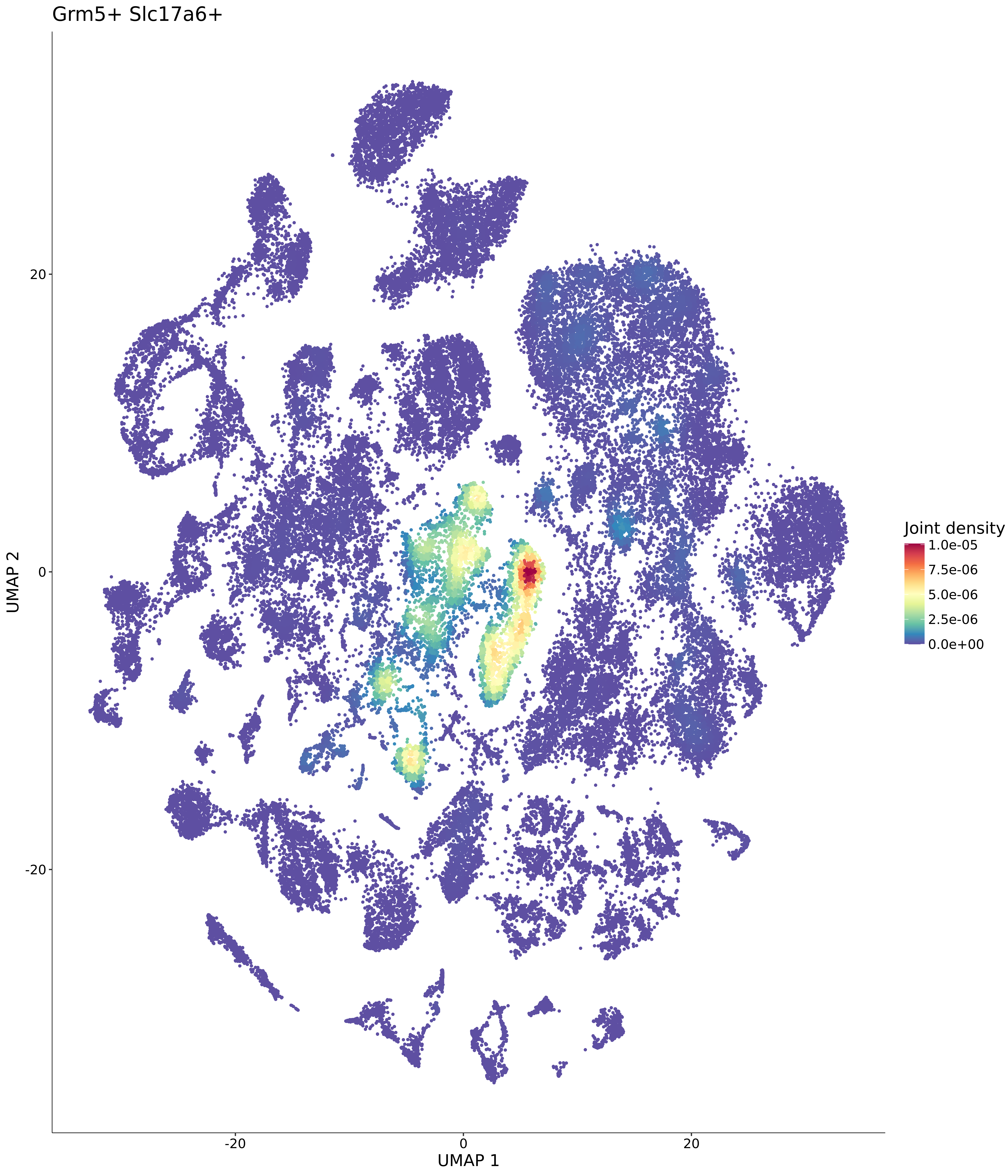
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Grm5" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
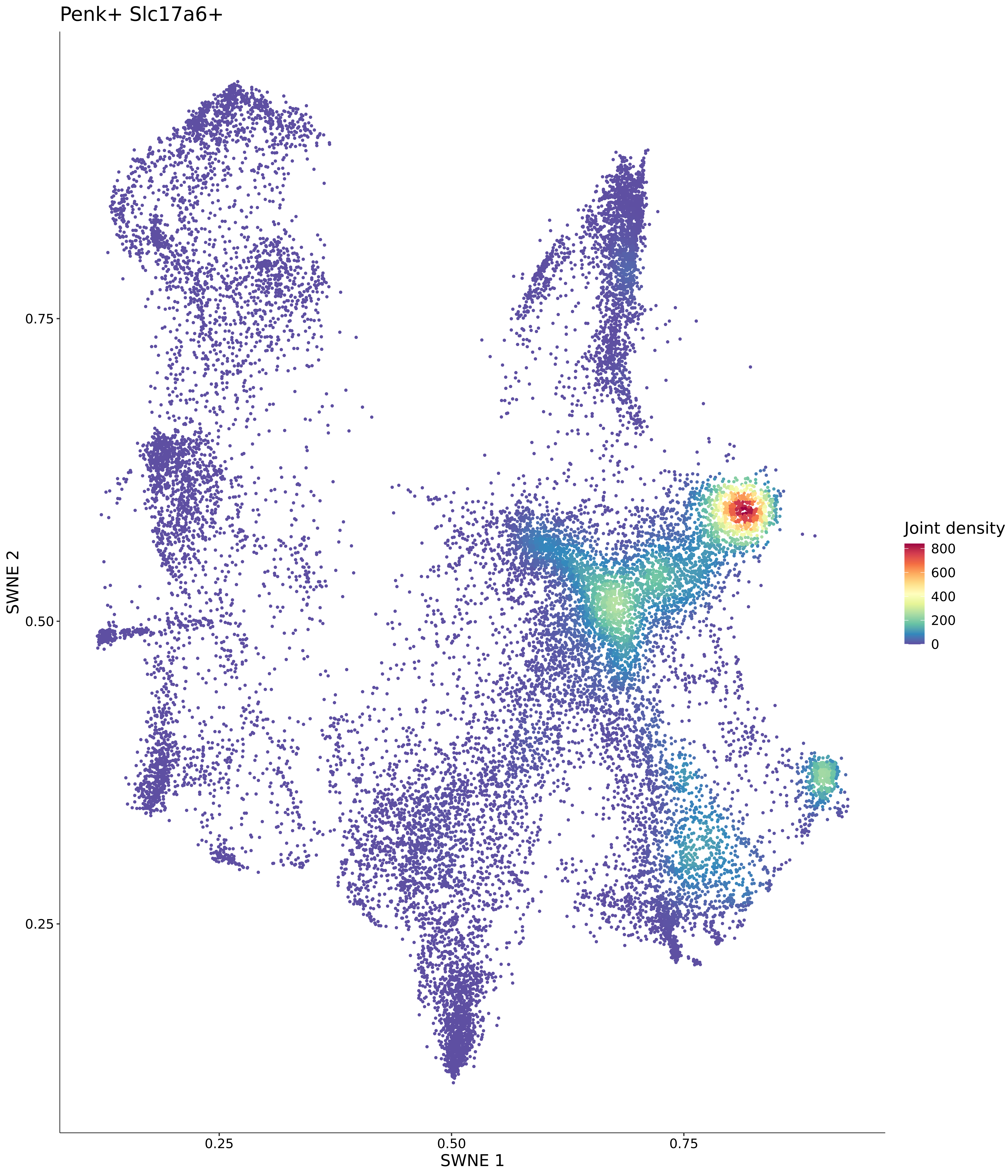
Code
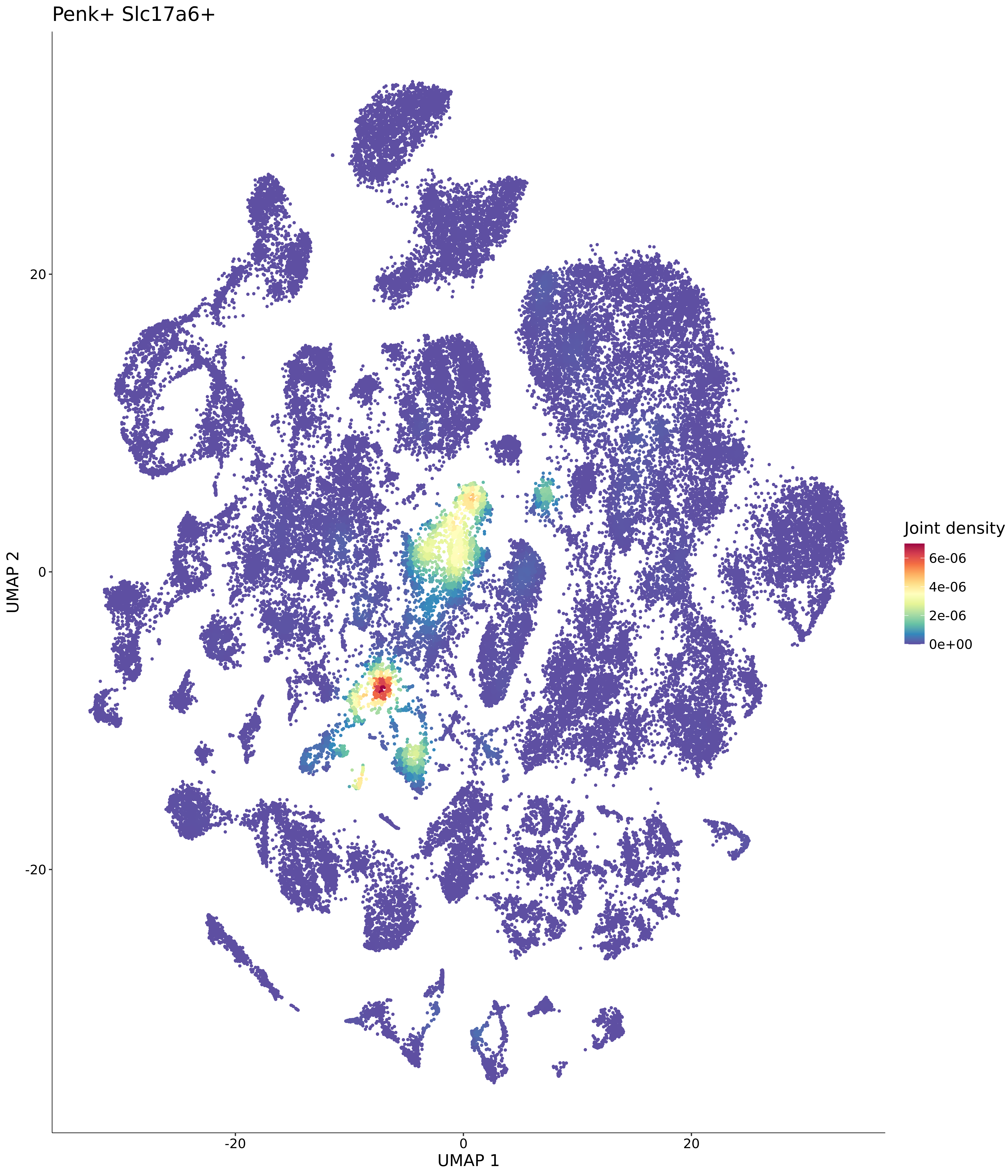
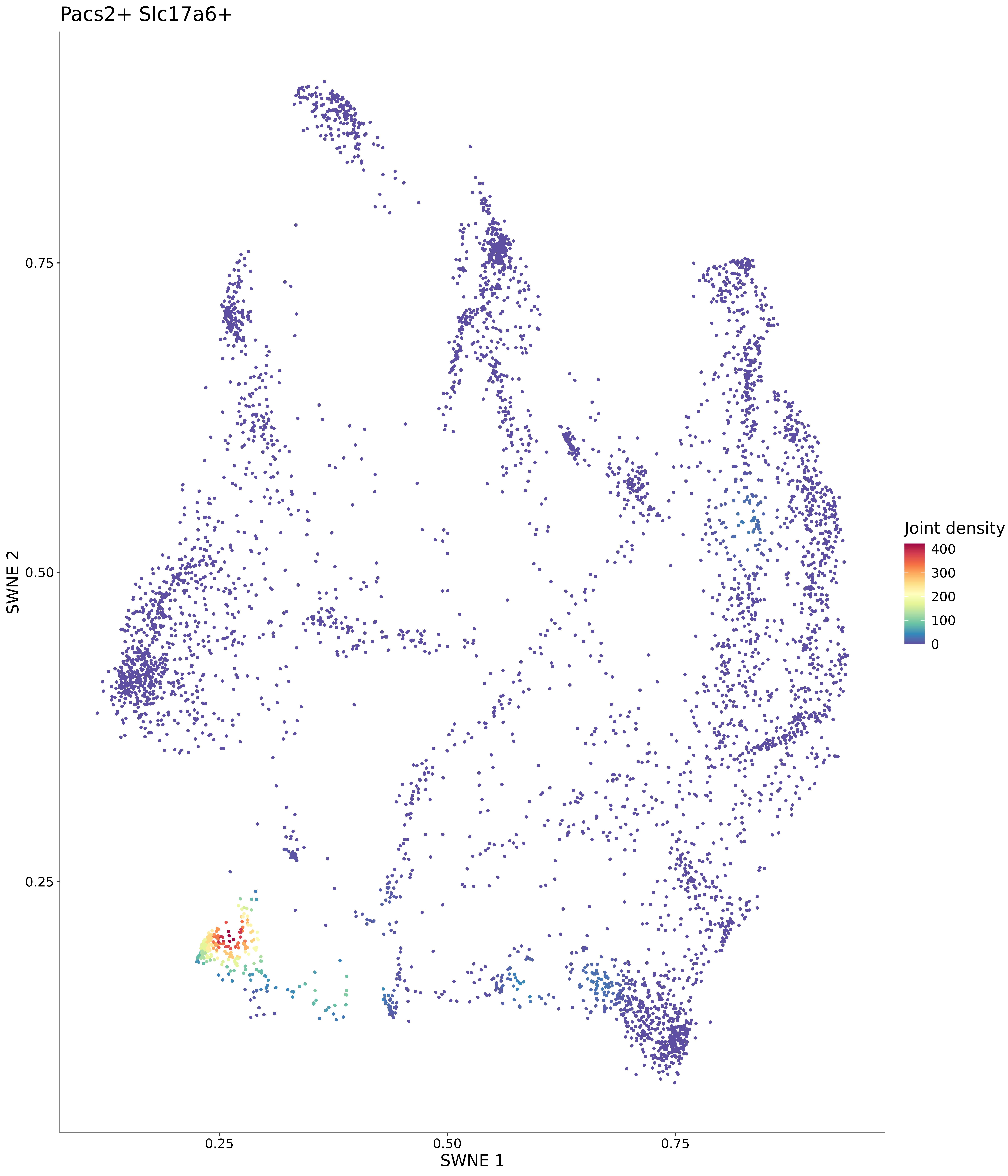
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Penk" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
Code
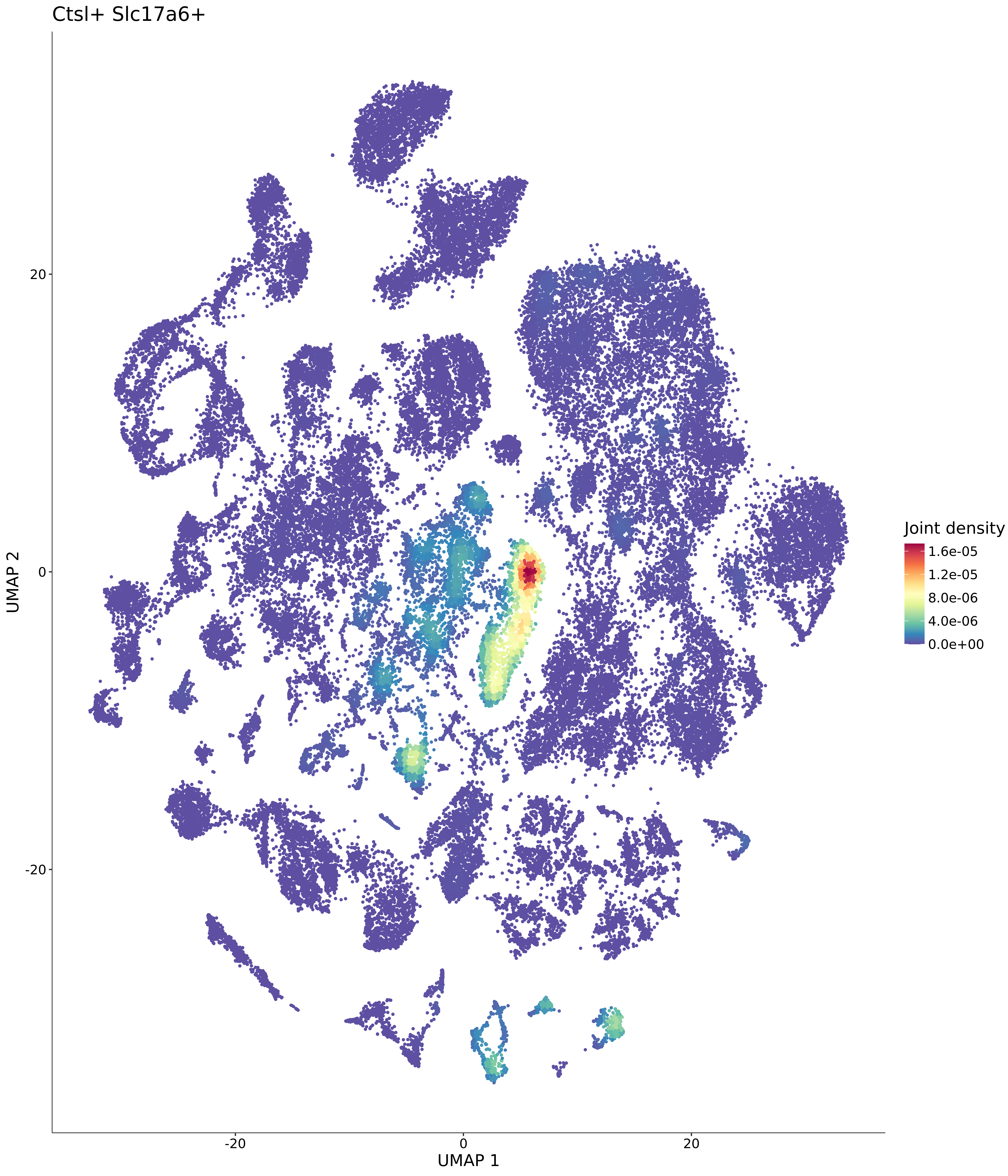
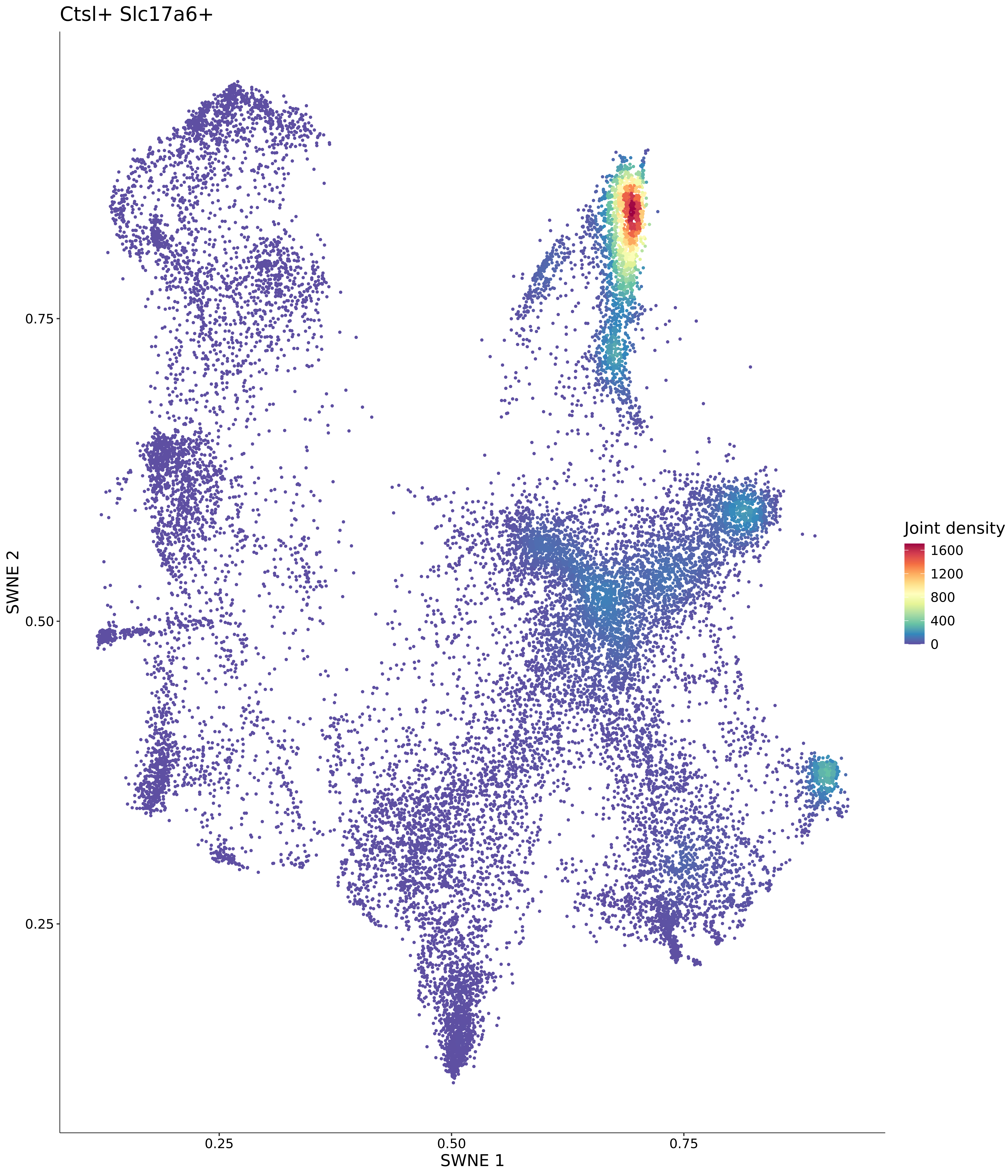
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Ctsl" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal)
Focused data analysis of Wdr37+ brain regions
Subset CA1 only
Code
<- subset (l6.srt, idents = "CA1" )<- FindVariableFeatures (ca1.srt, selection.method = "vst" , nfeatures = 3500 )<- ScaleData (ca1.srt)<- subset (l6.srt, idents = c ("CA1" , "Hypoth" , "MBd" , "MBv" , "Medulla" , "Pons" , "Thal" ))<- FindVariableFeatures (l6.srt, selection.method = "vst" , nfeatures = 3500 )<- ScaleData (l6.srt)<- VariableFeatures (l6.srt)<- Idents (l6.srt)
Run SWNE for CA1
Code
## Run SWNE <- RunSWNE (l6.srt,k = 20 , genes.embed = goi,return.format = "seurat"
calculating variance fit ... using gam [1] "3500 variable genes to use"
Initial stress : 0.23280
stress after 10 iters: 0.06662, magic = 0.500
stress after 20 iters: 0.06507, magic = 0.500
stress after 30 iters: 0.06492, magic = 0.500
stress after 40 iters: 0.06489, magic = 0.500
Code
<- RunSWNE (ca1.srt,k = 20 , genes.embed = goi,return.format = "seurat"
calculating variance fit ... using gam [1] "3500 variable genes to use"
Initial stress : 0.23150
stress after 10 iters: 0.07290, magic = 0.500
stress after 20 iters: 0.07265, magic = 0.500
stress after 30 iters: 0.07254, magic = 0.500
stress after 40 iters: 0.07251, magic = 0.500
Plot SWNE of CA1
Code
DimPlot_scCustom (reduction = "swne" ,pt.size = 0.5 ,group.by = "ClusterName" ,label = TRUE ,repel = TRUE ,figure_plot = TRUE ,color_seed = reseed& NoLegend ()
GABA
Code
Plot_Density_Custom (seurat_object = ca1.srt,features = c ("Slc32a1" ),custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne" )
Code
Plot_Density_Custom (seurat_object = ca1.srt,features = c ("Gad1" ),custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne" )
Code
Plot_Density_Custom (seurat_object = ca1.srt,features = c ("Gad2" ),custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne" )
Glutamate
Code
Plot_Density_Custom (seurat_object = ca1.srt,features = c ("Slc17a6" ),custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne" )
SWNE density plots of interactions between Wdr37 and other features
Code
Plot_Density_Joint_Only (seurat_object = ca1.srt, features = c ("Wdr37" , "Pacs2" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = ca1.srt, features = c ("Wdr37" , "Grm5" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = ca1.srt, features = c ("Grm5" , "Pacs2" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = ca1.srt, features = c ("Wdr37" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = ca1.srt, features = c ("Pacs2" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = ca1.srt, features = c ("Grm5" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Plot SWNE of selected regions: CA1, Hypothalamus, Midbrain, Medulla, Pons, Thalamus
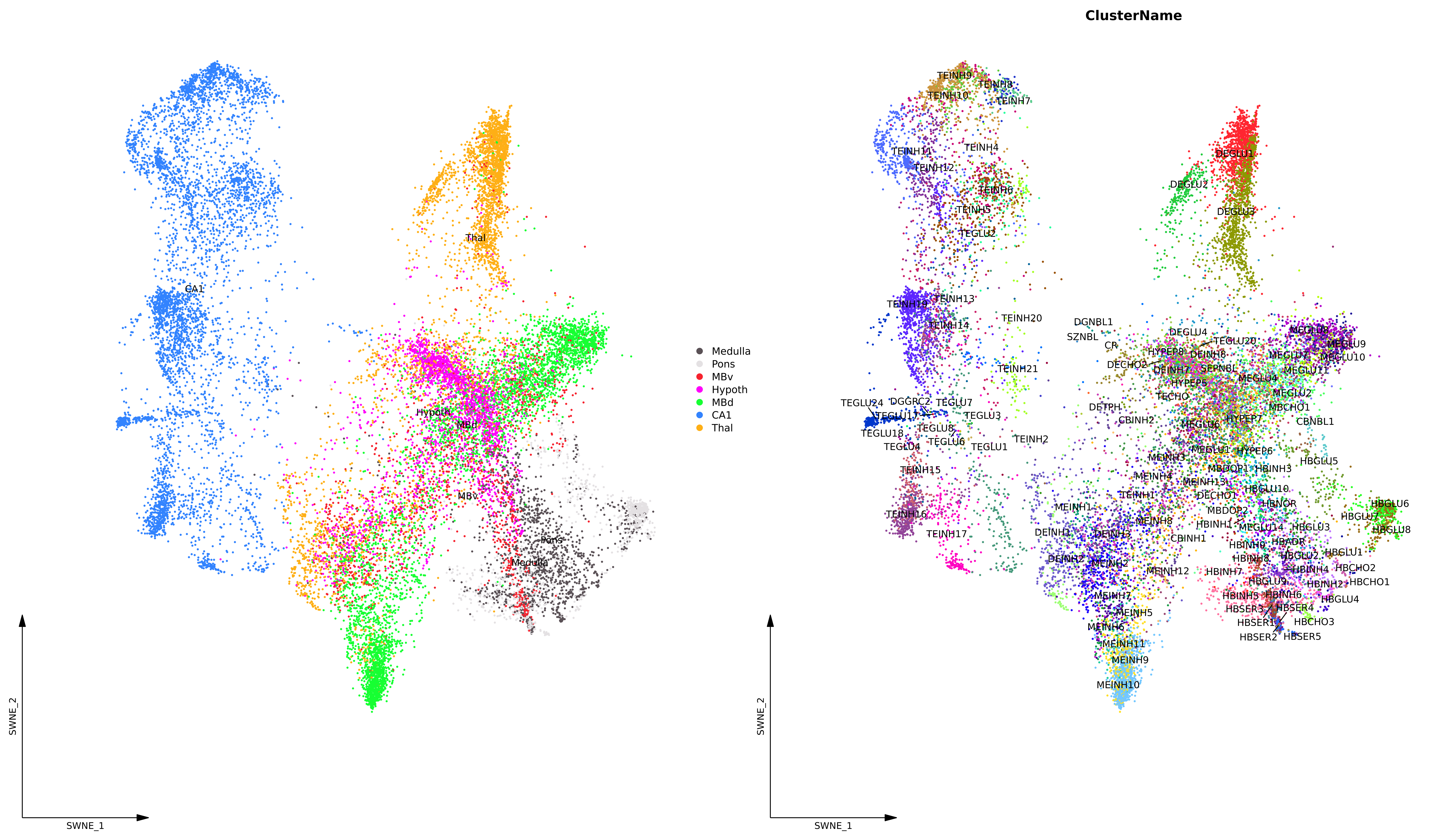
Code
DimPlot_scCustom (reduction = "swne" ,pt.size = 0.5 ,label = TRUE ,repel = TRUE ,figure_plot = TRUE ,color_seed = reseed| DimPlot_scCustom (reduction = "swne" ,pt.size = 0.5 ,group.by = "ClusterName" ,label = TRUE ,repel = TRUE ,figure_plot = TRUE ,color_seed = reseed& NoLegend ()
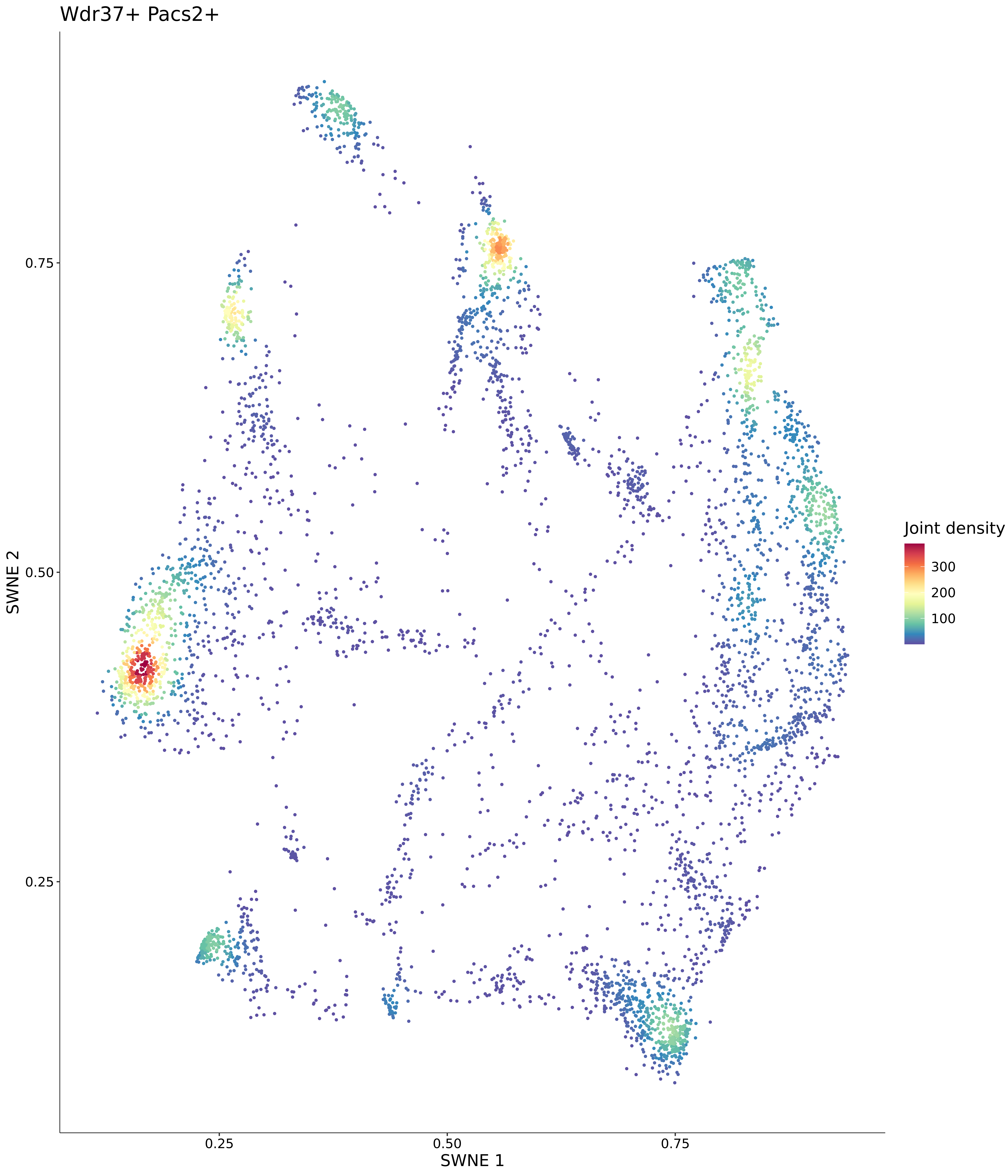
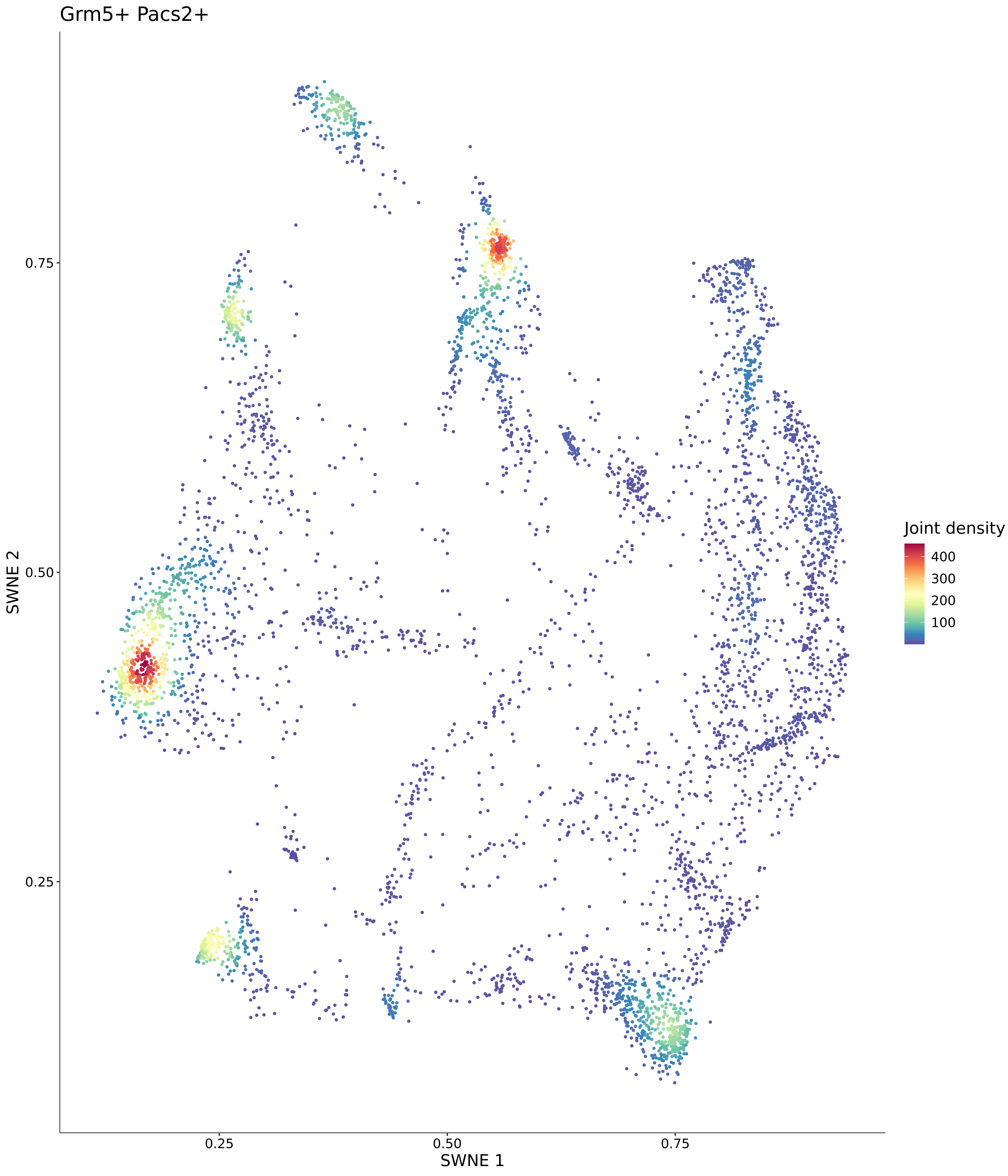
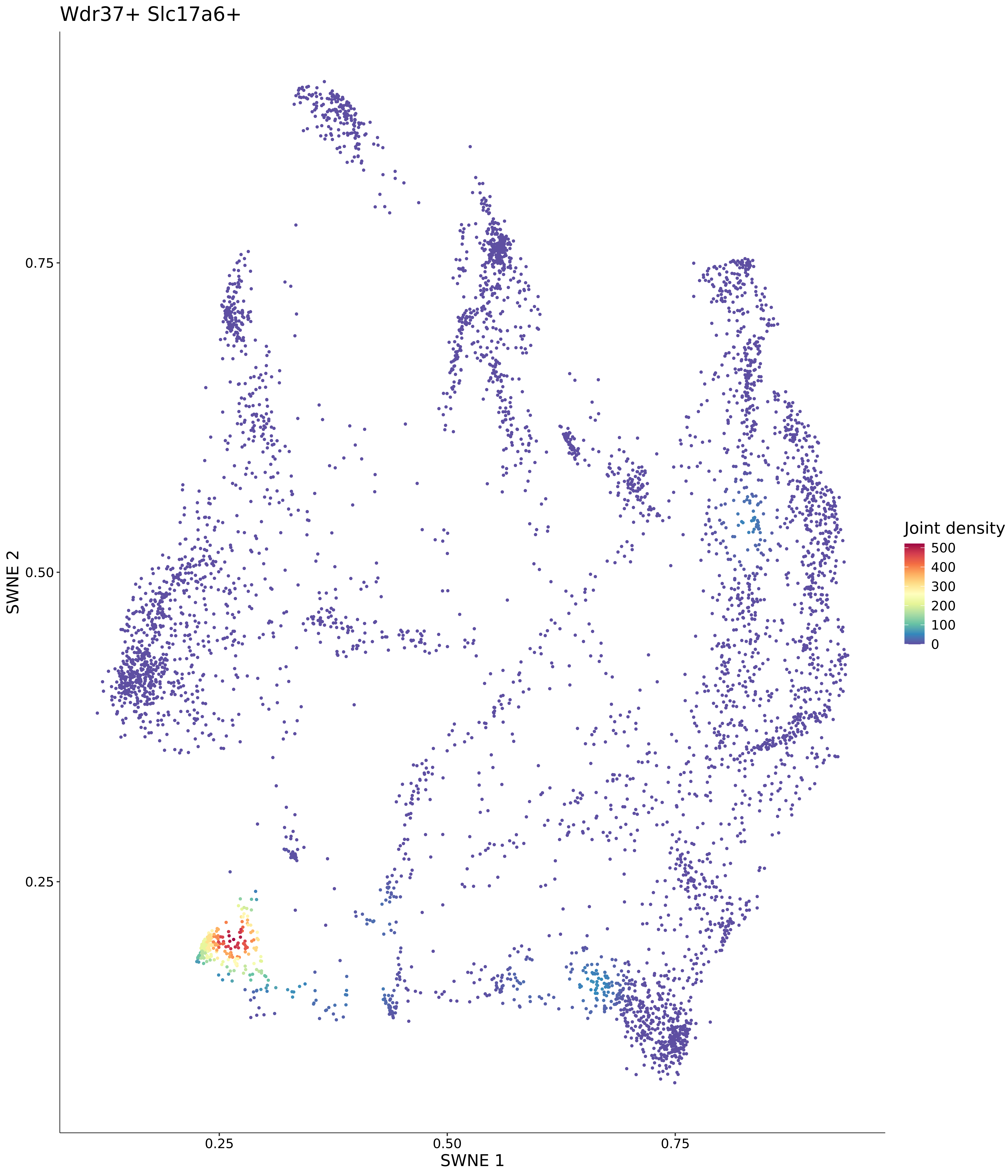
SWNE density plots of interactions between Wdr37 and other features
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Wdr37" , "Pacs2" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Wdr37" , "Grm5" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Grm5" , "Pacs2" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Wdr37" , "Penk" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
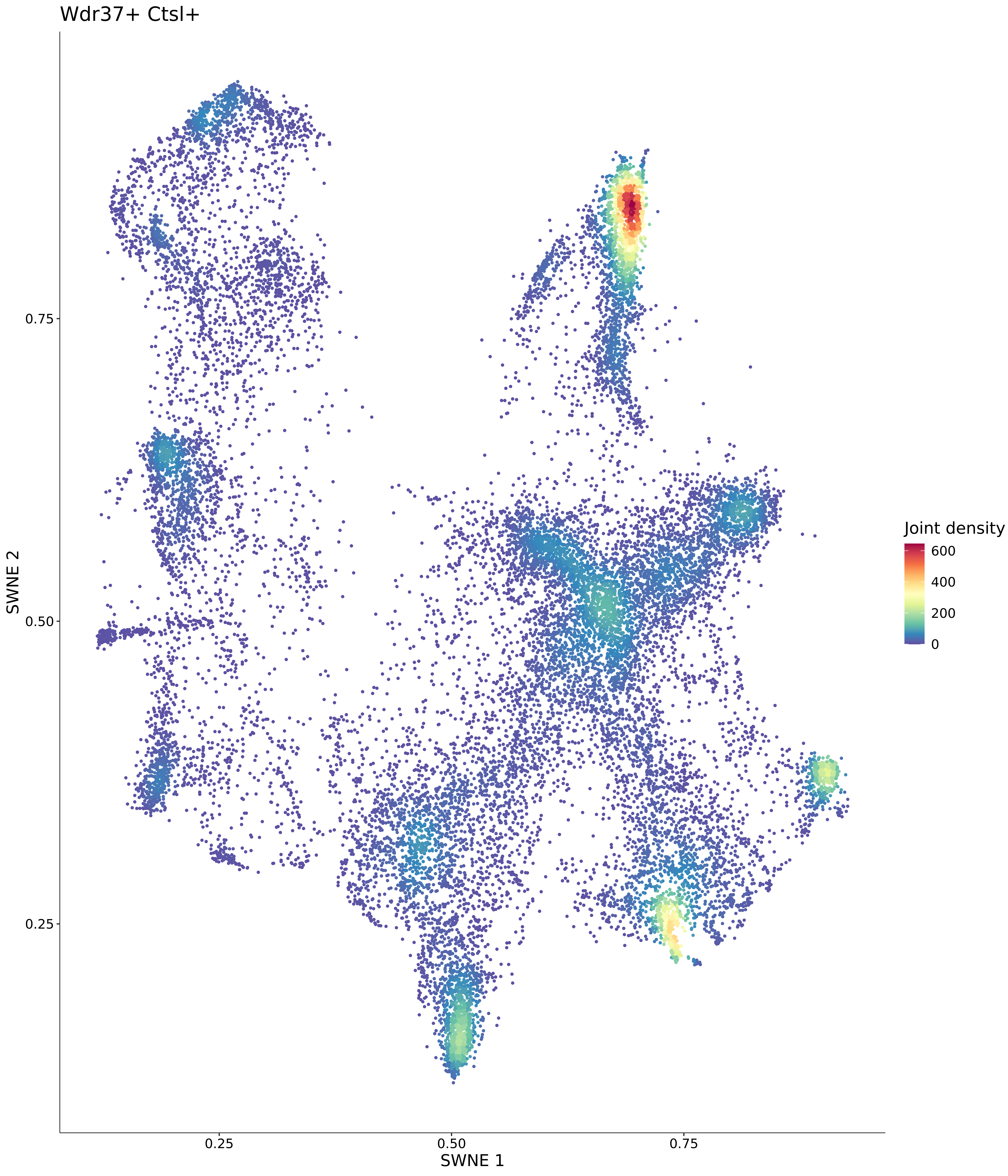
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Wdr37" , "Ctsl" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Wdr37" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Pacs2" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Grm5" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Penk" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Code
Plot_Density_Joint_Only (seurat_object = l6.srt, features = c ("Ctsl" , "Slc17a6" ), custom_palette = l6.srt@ misc$ div_Colour_Pal,reduction = "swne"
Calculate hexagon cell representation to quantify interactions between Wdr37 and other features in the SWNE space
Code
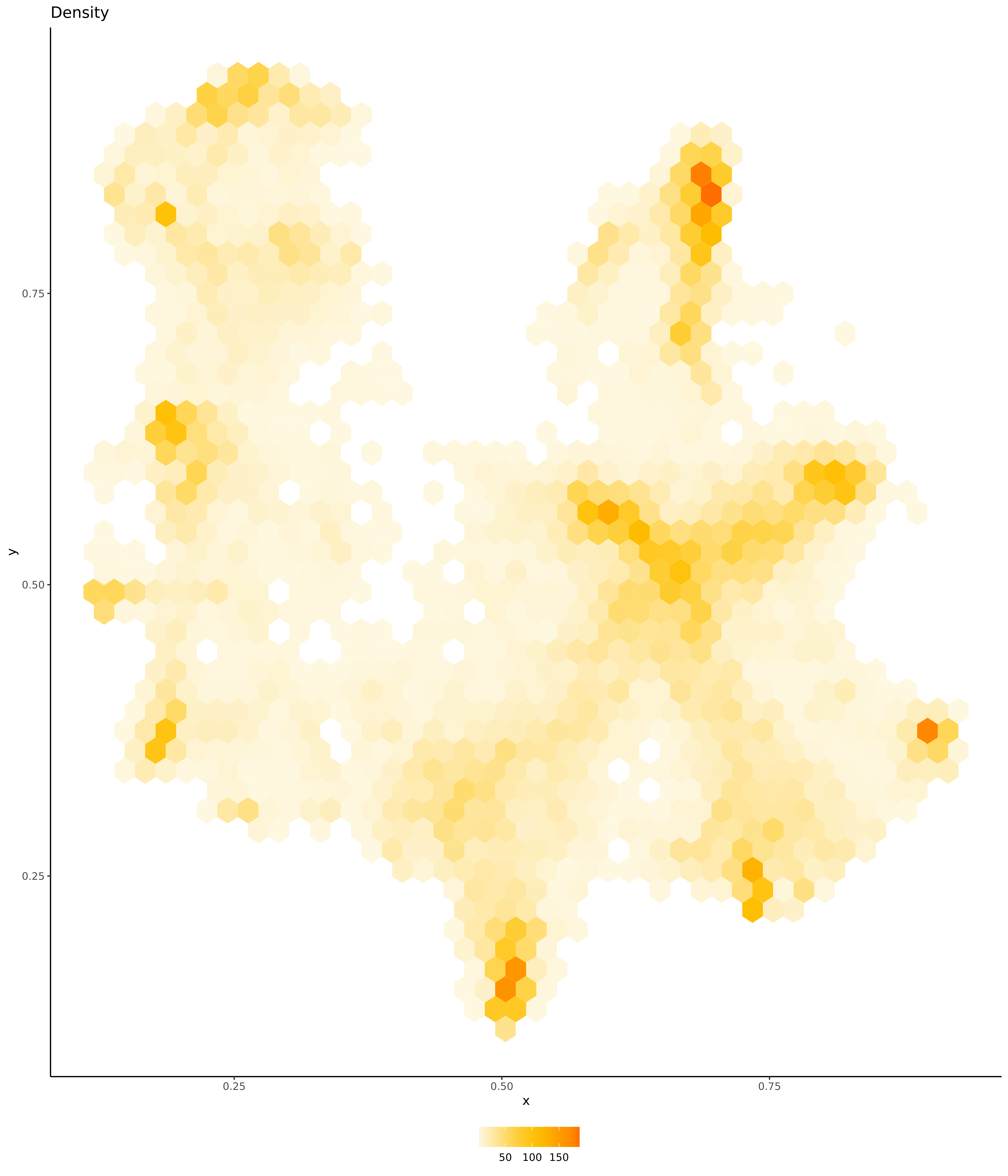
<- make_hexbin (l6.srt,nbins = 42 ,dimension_reduction = "swne" , use_dims = c (1 , 2 )
Plot hexagon cell representation
Code
plot_hexbin_density (l6.srt) + ggsci:: scale_fill_material ("amber" )
Code
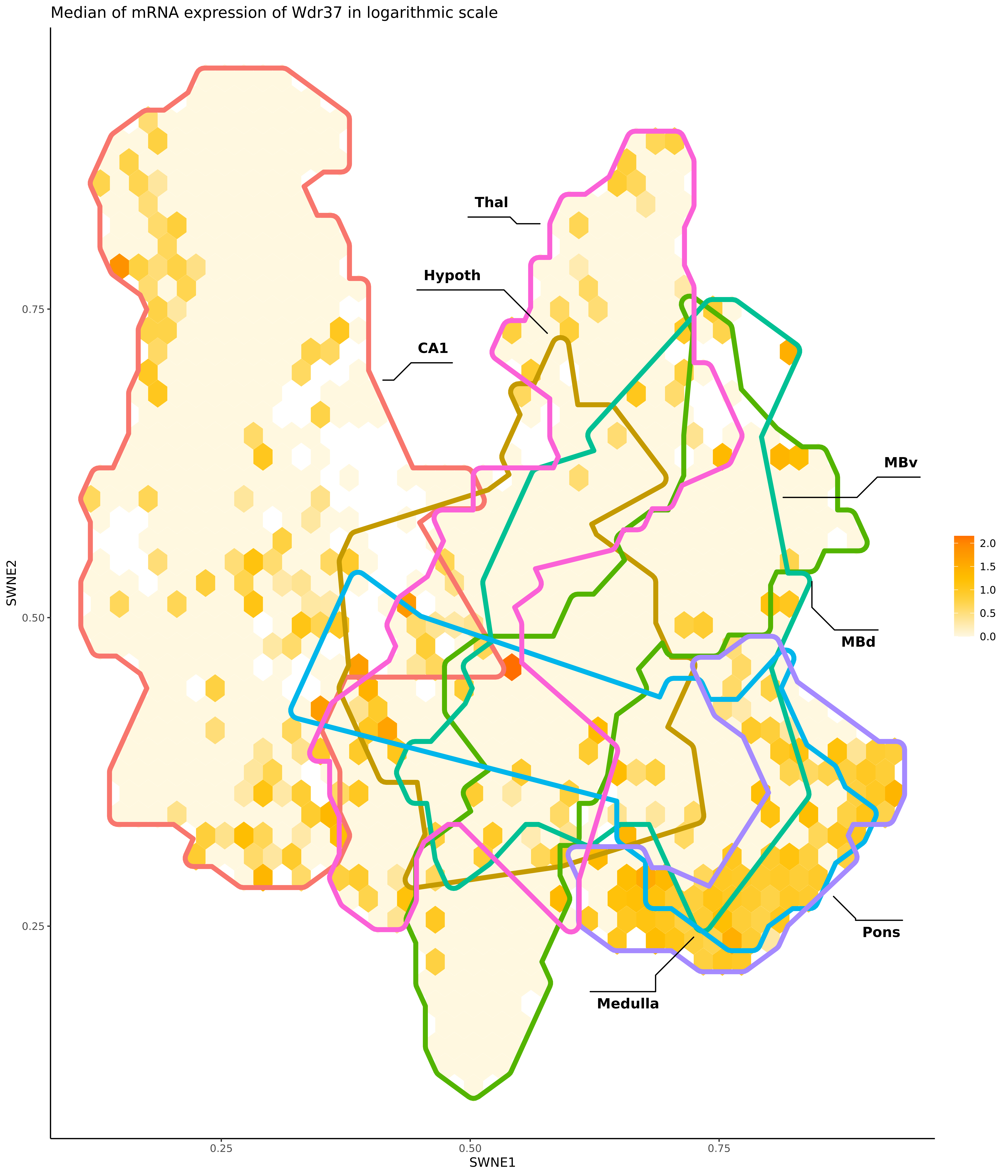
plot_hexbin_feature_plus (l6.srt,col = "Tissue" ,mod = "RNA" , type = "data" , feature = "Wdr37" ,action = "median" , xlab = "SWNE1" , ylab = "SWNE2" ,title = paste0 ("Median of mRNA expression of Wdr37 in logarithmic scale" )+ ggsci:: scale_fill_material ("amber" )
Code
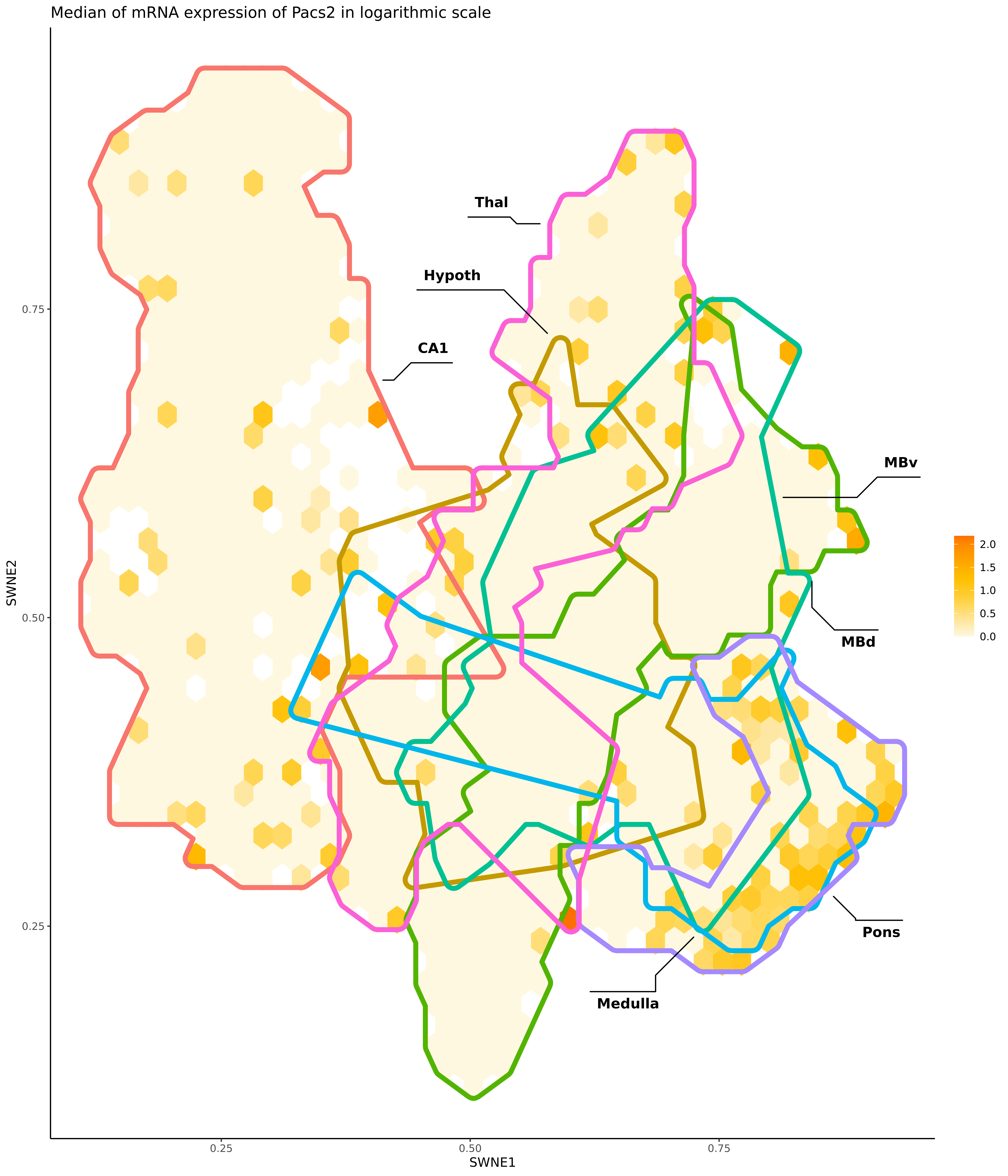
plot_hexbin_feature_plus (l6.srt,col = "Tissue" ,mod = "RNA" , type = "data" , feature = "Pacs2" ,action = "median" , xlab = "SWNE1" , ylab = "SWNE2" ,title = paste0 ("Median of mRNA expression of Pacs2 in logarithmic scale" )+ ggsci:: scale_fill_material ("amber" )
Code
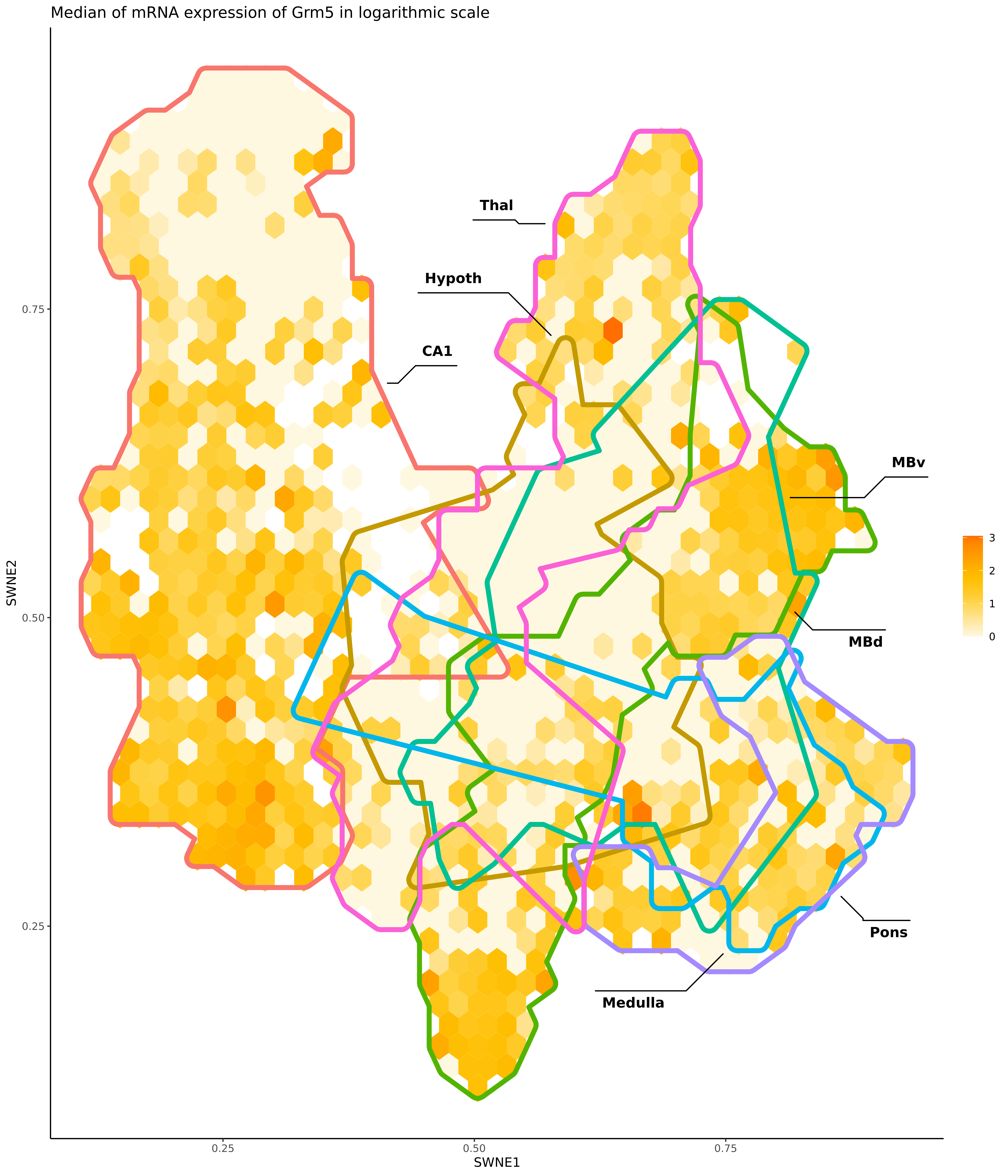
plot_hexbin_feature_plus (l6.srt,col = "Tissue" ,mod = "RNA" , type = "data" , feature = "Grm5" ,action = "median" , xlab = "SWNE1" , ylab = "SWNE2" ,title = paste0 ("Median of mRNA expression of Grm5 in logarithmic scale" )+ ggsci:: scale_fill_material ("amber" )
Code
plot_hexbin_feature_plus (l6.srt,col = "Tissue" ,mod = "RNA" , type = "data" , feature = "Slc32a1" ,action = "median" , xlab = "SWNE1" , ylab = "SWNE2" ,title = paste0 ("Median of mRNA expression of Slc32a1 in logarithmic scale" )+ ggsci:: scale_fill_material ("amber" )
Code
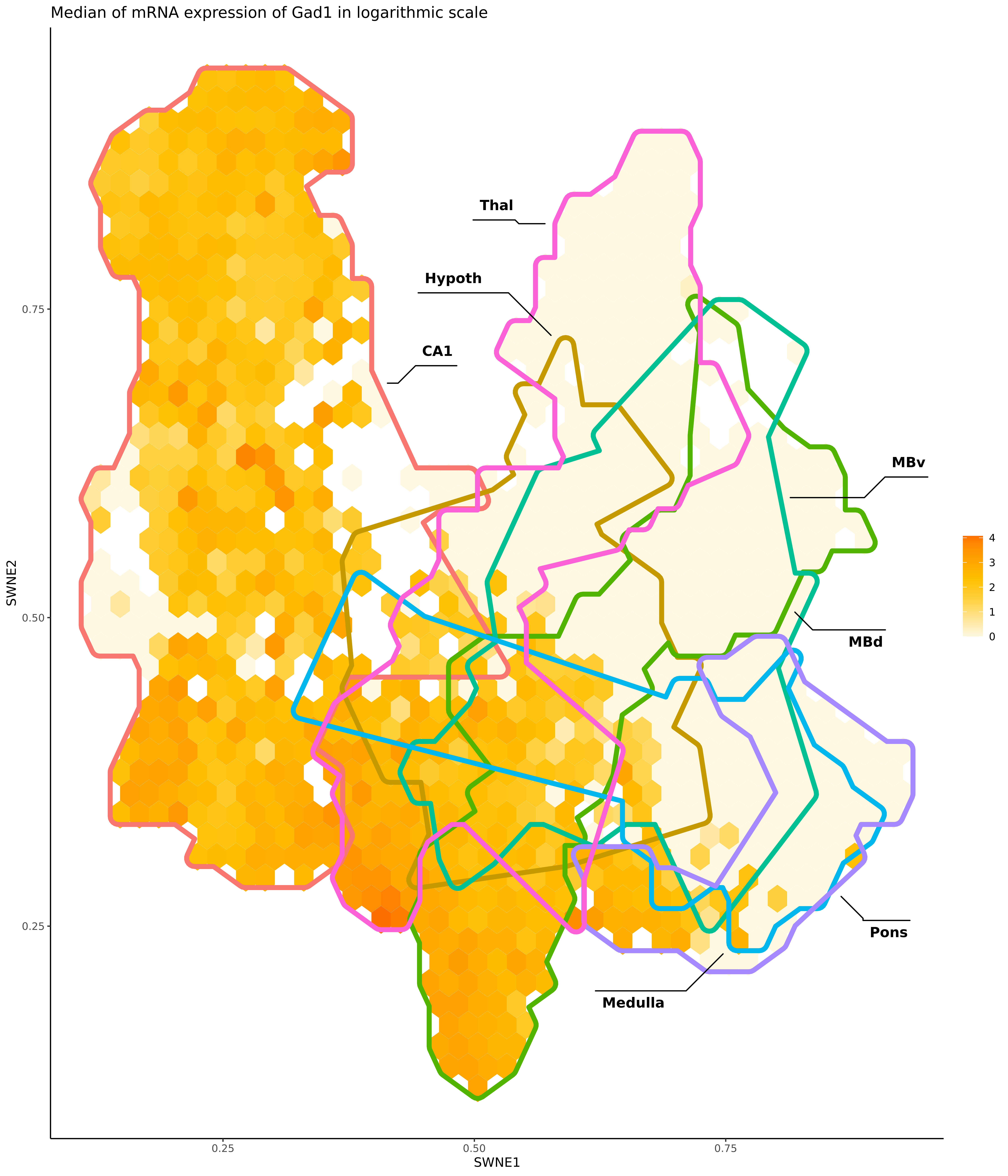
plot_hexbin_feature_plus (l6.srt,col = "Tissue" ,mod = "RNA" , type = "data" , feature = "Gad1" ,action = "median" , xlab = "SWNE1" , ylab = "SWNE2" ,title = paste0 ("Median of mRNA expression of Gad1 in logarithmic scale" )+ ggsci:: scale_fill_material ("amber" )
Code
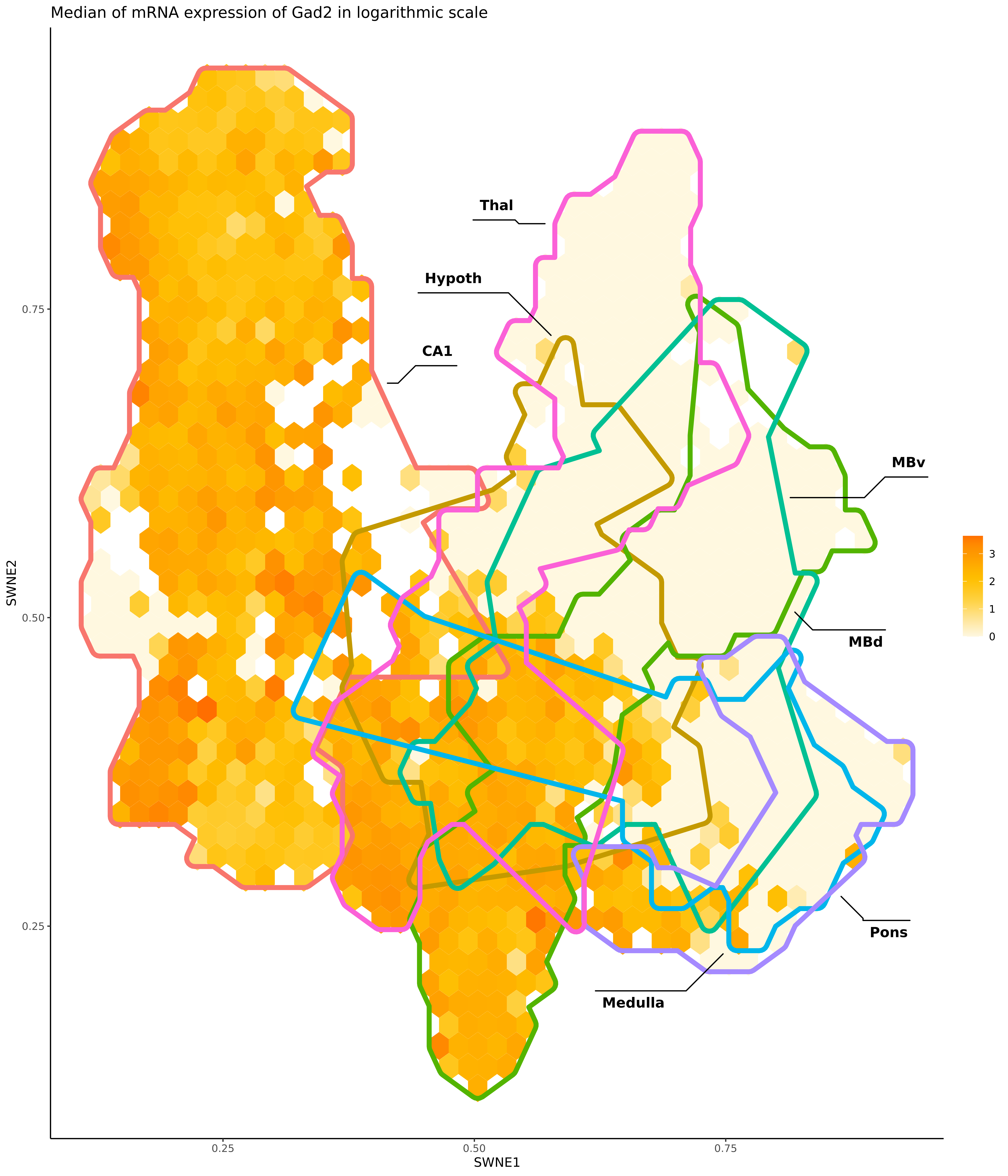
plot_hexbin_feature_plus (l6.srt,col = "Tissue" ,mod = "RNA" , type = "data" , feature = "Gad2" ,action = "median" , xlab = "SWNE1" , ylab = "SWNE2" ,title = paste0 ("Median of mRNA expression of Gad2 in logarithmic scale" )+ ggsci:: scale_fill_material ("amber" )
Code
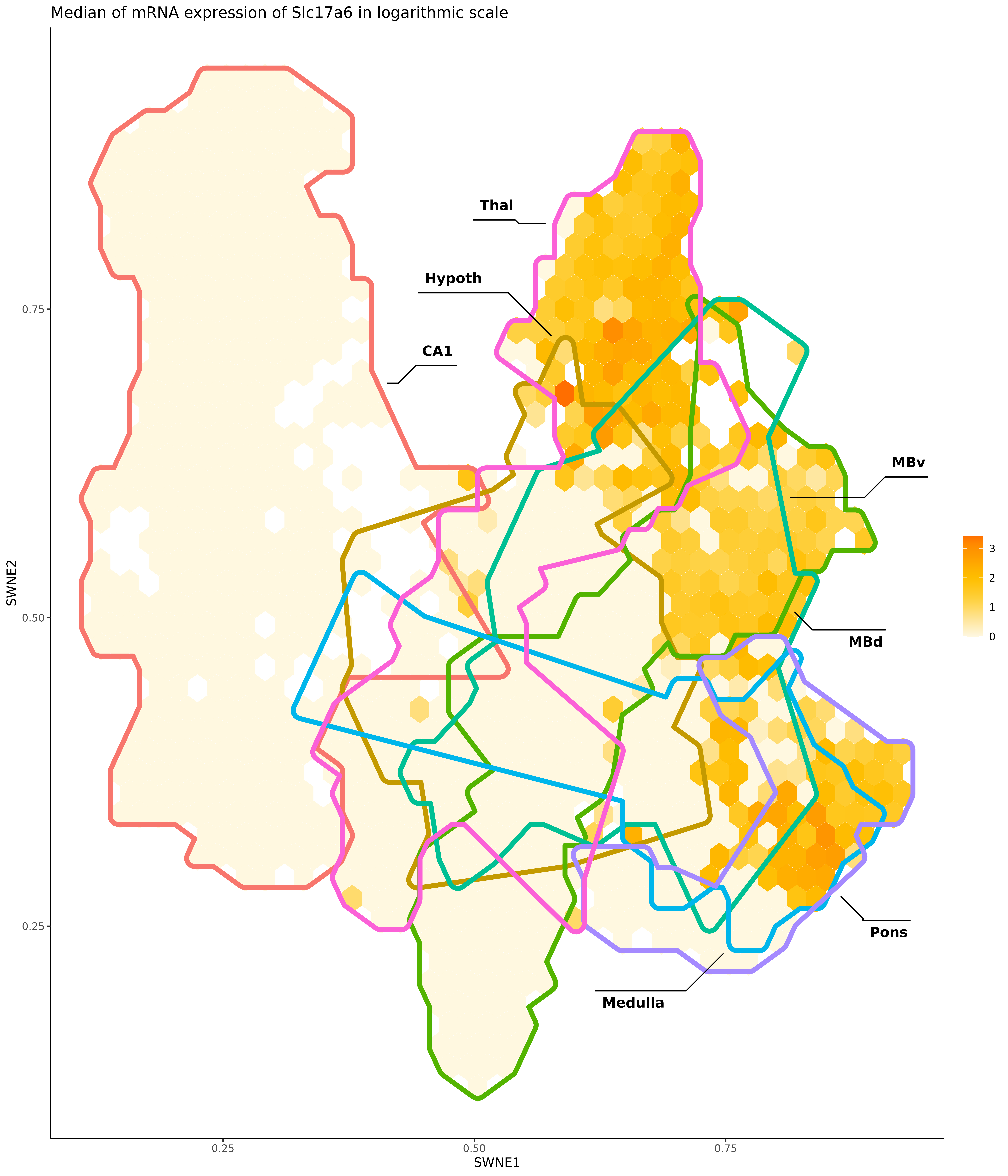
plot_hexbin_feature_plus (l6.srt,col = "Tissue" ,mod = "RNA" , type = "data" , feature = "Slc17a6" ,action = "median" , xlab = "SWNE1" , ylab = "SWNE2" ,title = paste0 ("Median of mRNA expression of Slc17a6 in logarithmic scale" )+ ggsci:: scale_fill_material ("amber" )
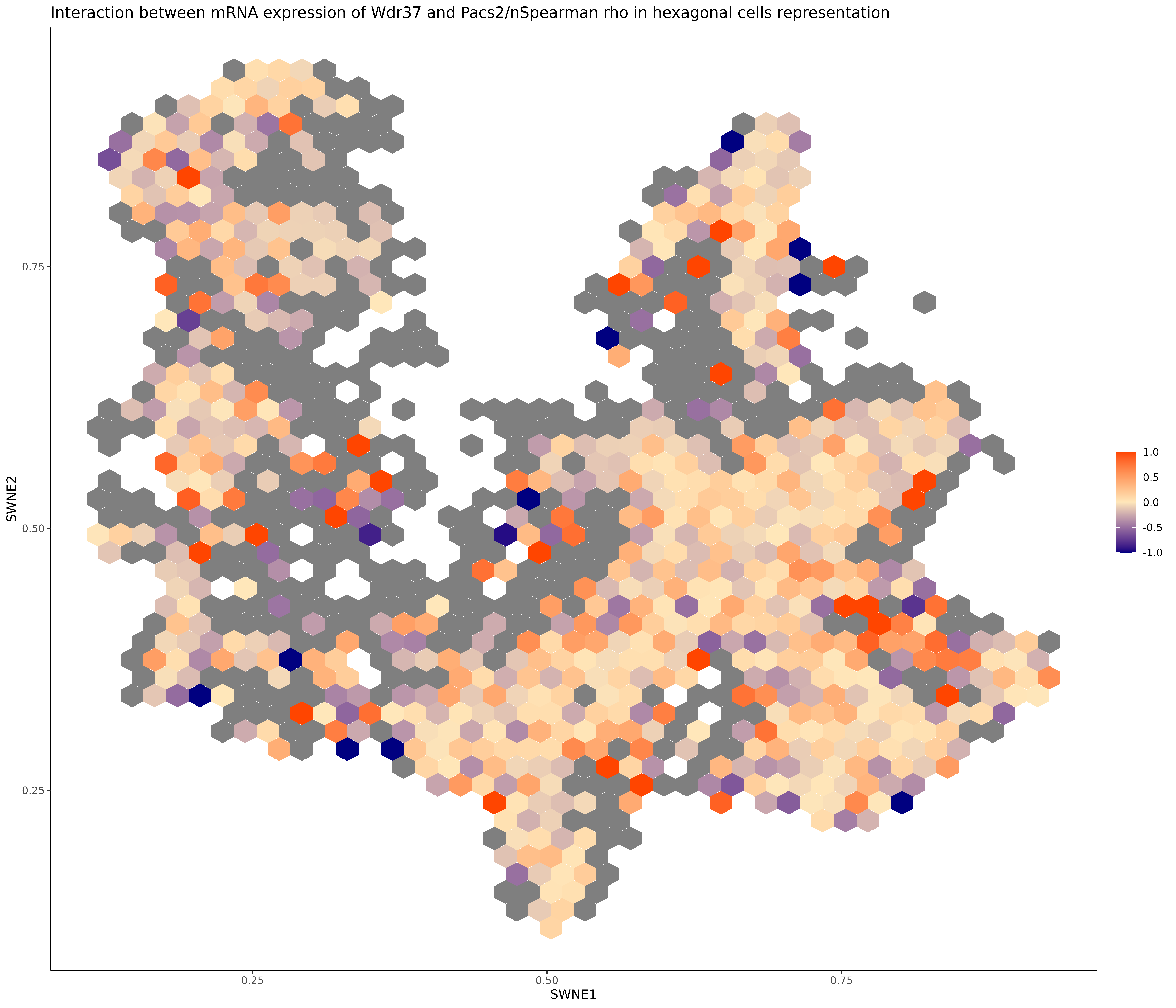
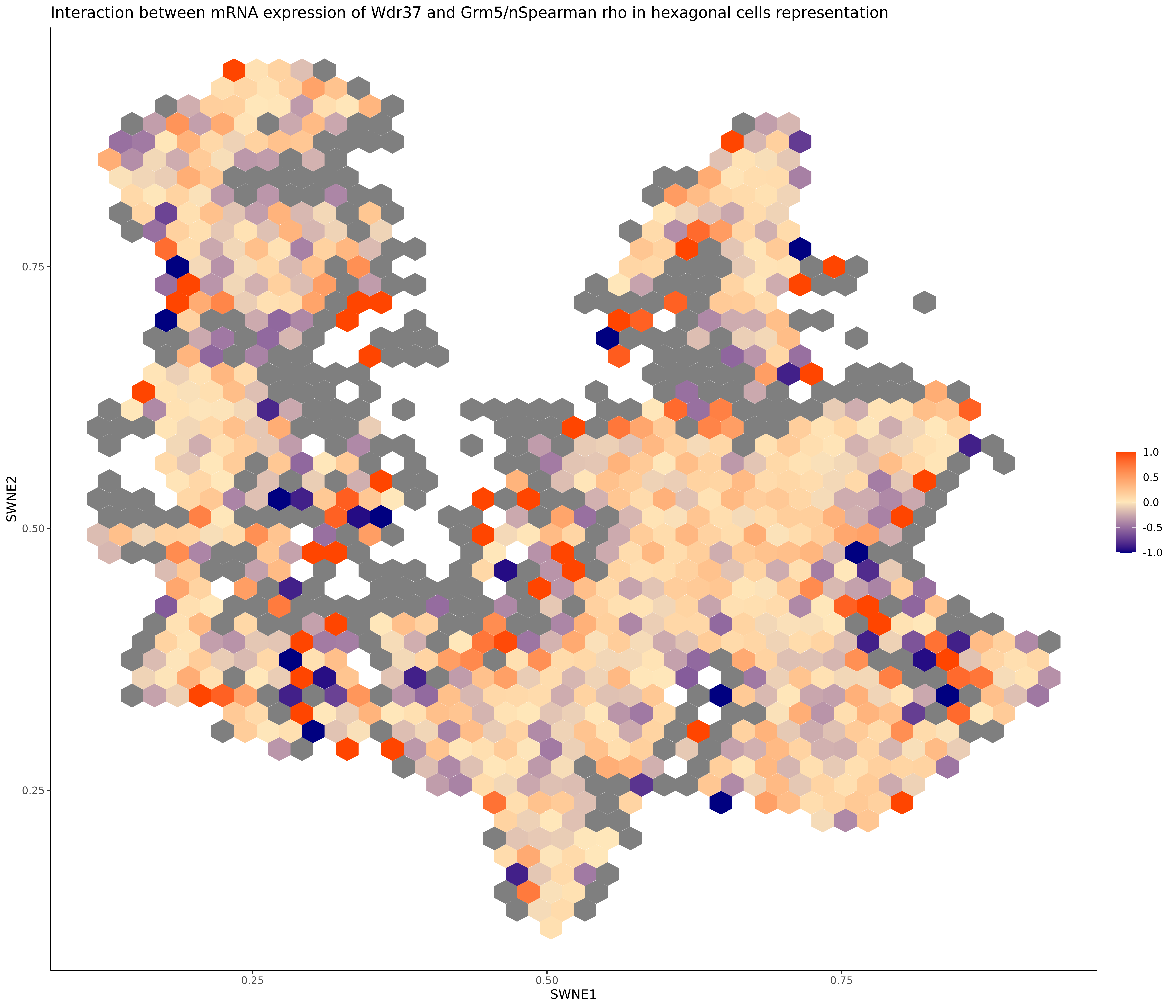
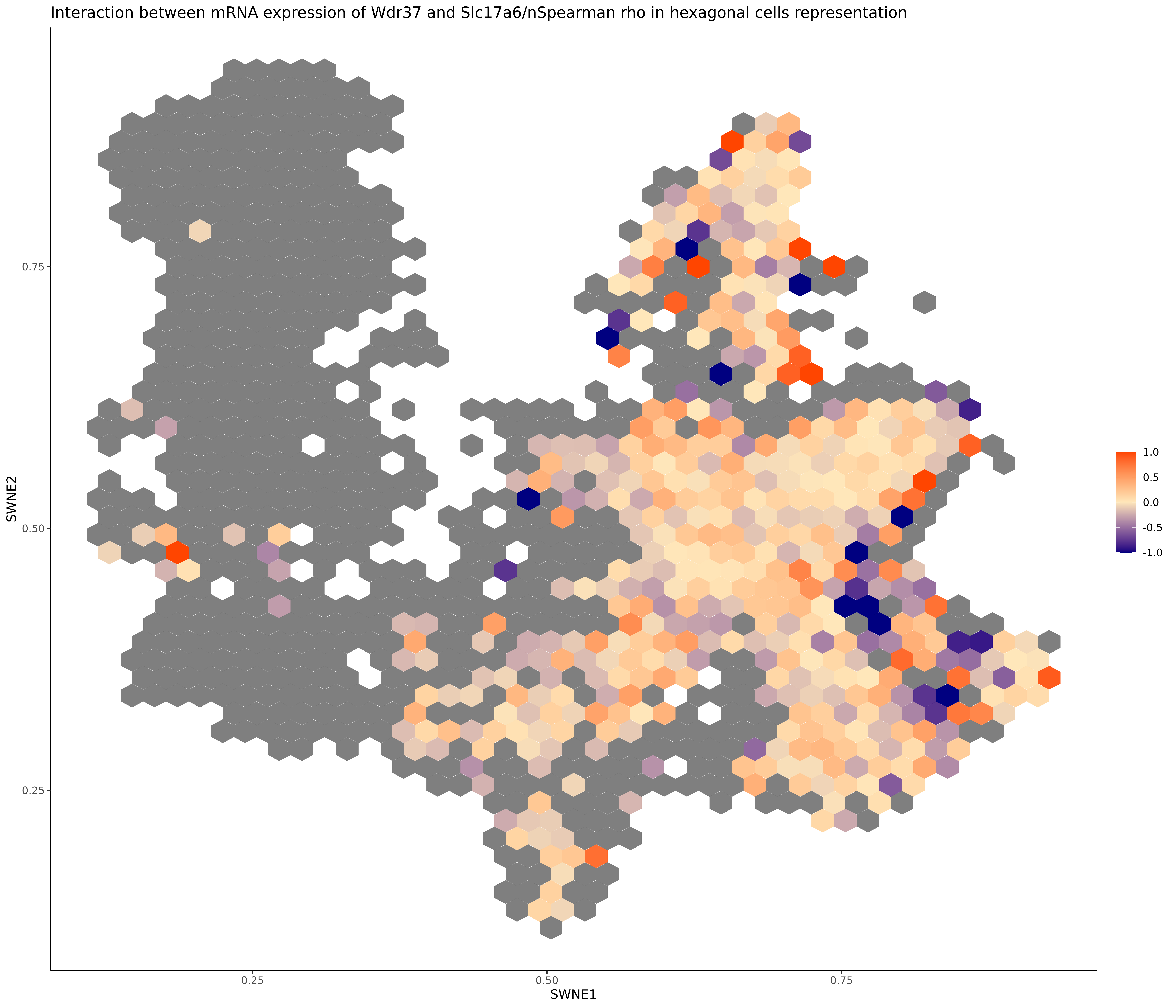
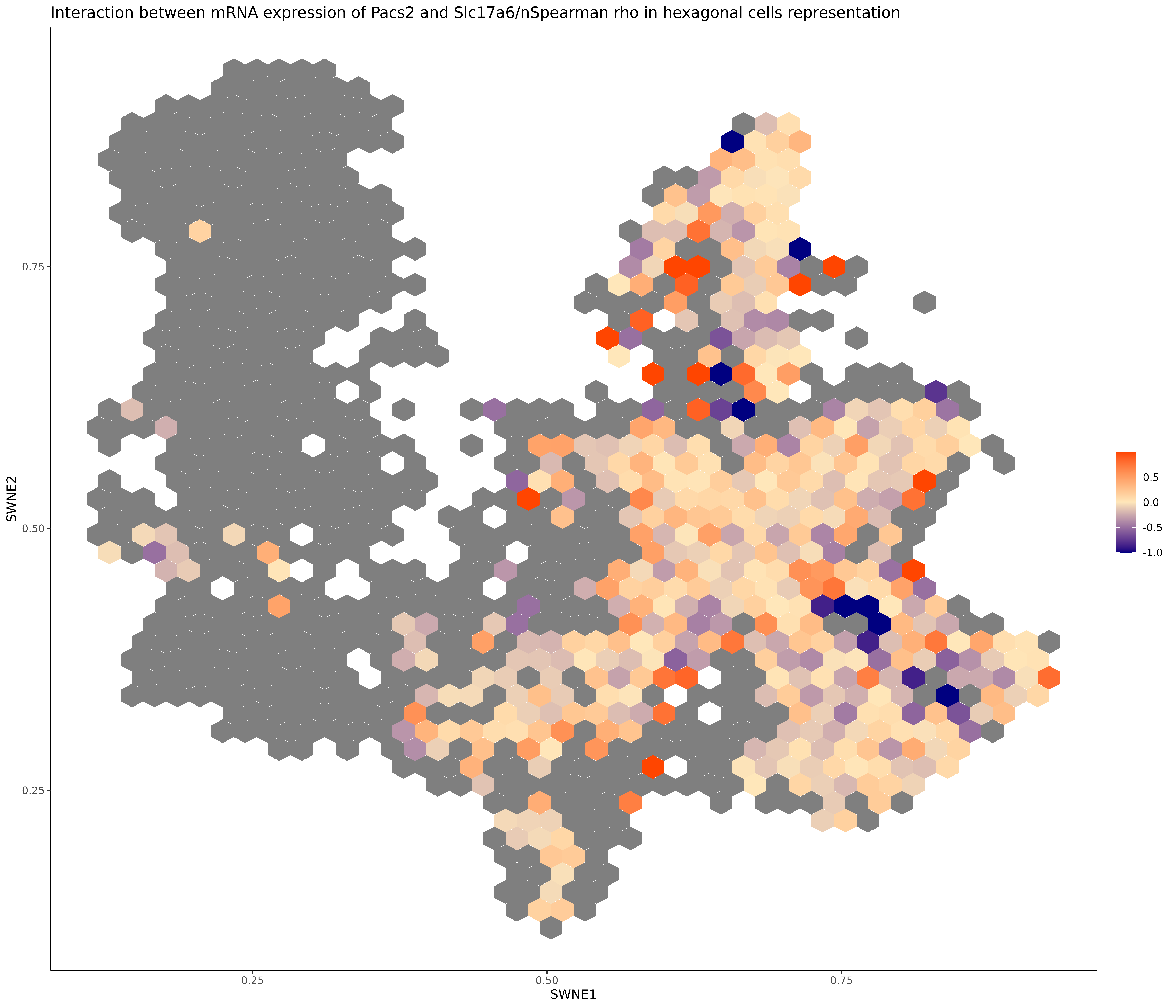
Quantify and plot hexagon representation of feature interactions with Spearman’s correlation
Code
plot_hexbin_interact (l6.srt,type = c ("data" , "data" ),mod = c ("RNA" , "RNA" ), feature = c ("Wdr37" , "Pacs2" ), interact = "corr_spearman" ,ylab = "SWNE2" , xlab = "SWNE1" ,title = "Interaction between mRNA expression of Wdr37 and Pacs2/nSpearman rho in hexagonal cells representation" + scale_fill_gradient2 (midpoint = 0 , space = "Lab" ,low = "navy" ,mid = "wheat1" ,high = "orangered1"
Code
plot_hexbin_interact (l6.srt,type = c ("data" , "data" ),mod = c ("RNA" , "RNA" ), feature = c ("Wdr37" , "Grm5" ), interact = "corr_spearman" ,ylab = "SWNE2" , xlab = "SWNE1" ,title = "Interaction between mRNA expression of Wdr37 and Grm5/nSpearman rho in hexagonal cells representation" + scale_fill_gradient2 (midpoint = 0 , space = "Lab" ,low = "navy" ,mid = "wheat1" ,high = "orangered1"
Code
plot_hexbin_interact (l6.srt,type = c ("data" , "data" ),mod = c ("RNA" , "RNA" ), feature = c ("Wdr37" , "Slc17a6" ), interact = "corr_spearman" ,ylab = "SWNE2" , xlab = "SWNE1" ,title = "Interaction between mRNA expression of Wdr37 and Slc17a6/nSpearman rho in hexagonal cells representation" + scale_fill_gradient2 (midpoint = 0 , space = "Lab" ,low = "navy" ,mid = "wheat1" ,high = "orangered1"
Code
plot_hexbin_interact (l6.srt,type = c ("data" , "data" ),mod = c ("RNA" , "RNA" ), feature = c ("Pacs2" , "Slc17a6" ), interact = "corr_spearman" ,ylab = "SWNE2" , xlab = "SWNE1" ,title = "Interaction between mRNA expression of Pacs2 and Slc17a6/nSpearman rho in hexagonal cells representation" + scale_fill_gradient2 (midpoint = 0 , space = "Lab" ,low = "navy" ,mid = "wheat1" ,high = "orangered1"
Code
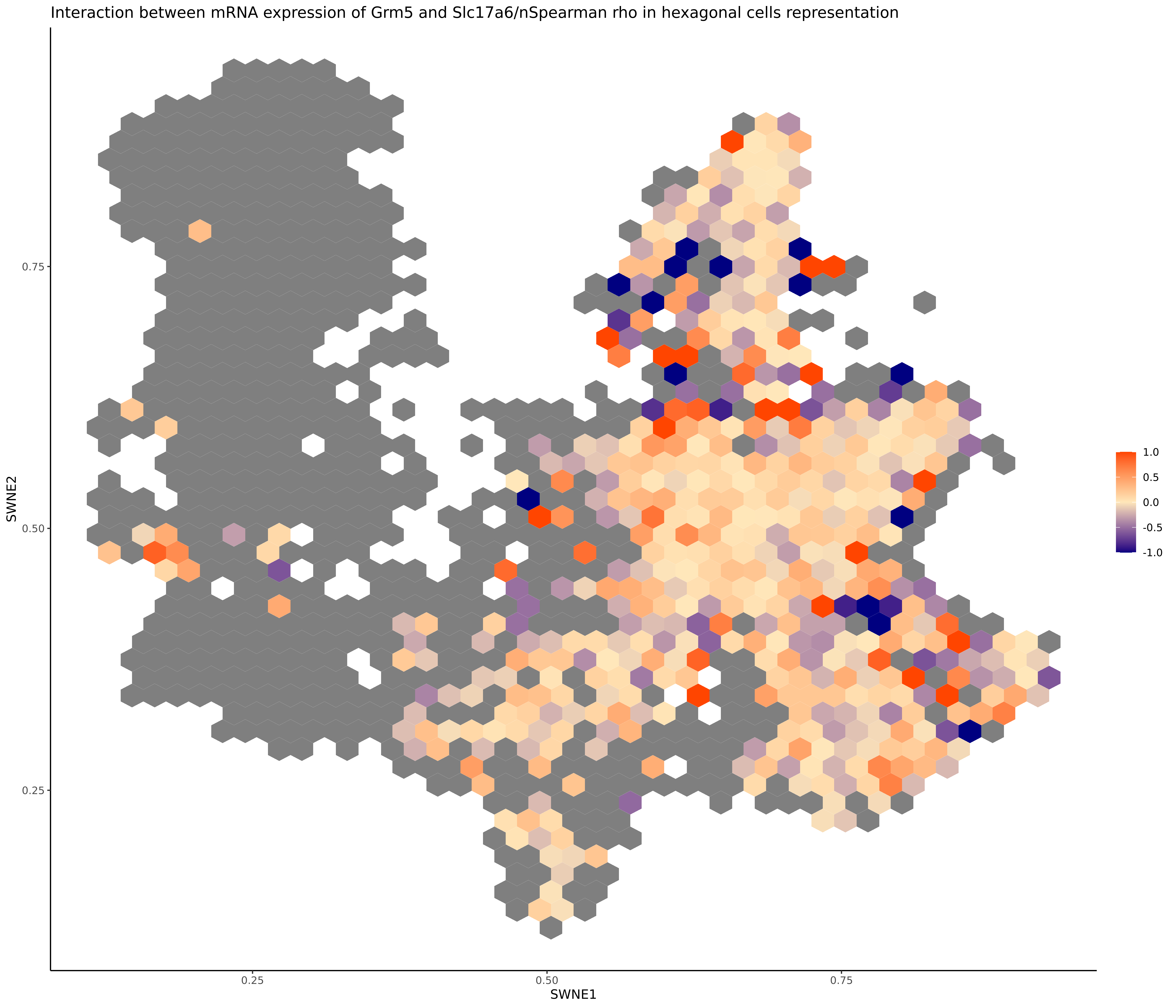
plot_hexbin_interact (l6.srt,type = c ("data" , "data" ),mod = c ("RNA" , "RNA" ), feature = c ("Grm5" , "Slc17a6" ), interact = "corr_spearman" ,ylab = "SWNE2" , xlab = "SWNE1" ,title = "Interaction between mRNA expression of Grm5 and Slc17a6/nSpearman rho in hexagonal cells representation" + scale_fill_gradient2 (midpoint = 0 , space = "Lab" ,low = "navy" ,mid = "wheat1" ,high = "orangered1"
Save results
Code
write_rds (l6.srt, file = here ("l6.srt.rds" ))
Code
:: session_info ()
─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.2.2 (2022-10-31)
os Ubuntu 22.04.2 LTS
system x86_64, linux-gnu
ui X11
language en_US:en
collate en_US.UTF-8
ctype en_US.UTF-8
tz Etc/UTC
date 2023-08-02
pandoc 2.19.2 @ /opt/python/3.8.8/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
abind 1.4-5 2016-07-21 [1] RSPM (R 4.2.0)
askpass 1.1 2019-01-13 [1] RSPM (R 4.2.0)
base64enc 0.1-3 2015-07-28 [1] RSPM (R 4.2.0)
beeswarm 0.4.0 2021-06-01 [1] RSPM (R 4.2.0)
Biobase * 2.58.0 2022-11-01 [1] RSPM (R 4.2.2)
BiocGenerics * 0.44.0 2022-11-01 [1] RSPM (R 4.2.2)
BiocManager 1.30.20 2023-02-24 [1] RSPM (R 4.2.0)
bit 4.0.5 2022-11-15 [1] RSPM (R 4.2.0)
bit64 4.0.5 2020-08-30 [1] RSPM (R 4.2.0)
bitops 1.0-7 2021-04-24 [1] RSPM (R 4.2.0)
circlize 0.4.15 2022-05-10 [1] RSPM (R 4.2.0)
cli 3.6.1 2023-03-23 [1] RSPM (R 4.2.0)
cluster 2.1.4 2022-08-22 [1] RSPM (R 4.2.0)
codetools 0.2-19 2023-02-01 [1] RSPM (R 4.2.0)
colorspace 2.1-0 2023-01-23 [1] RSPM (R 4.2.0)
concaveman 1.1.0 2020-05-11 [1] RSPM (R 4.2.0)
cowplot * 1.1.1 2020-12-30 [1] RSPM (R 4.2.0)
crayon 1.5.2 2022-09-29 [1] RSPM (R 4.2.0)
curl 5.0.0 2023-01-12 [1] RSPM (R 4.2.0)
data.table 1.14.8 2023-02-17 [1] RSPM (R 4.2.0)
DelayedArray 0.24.0 2022-11-01 [1] RSPM (R 4.2.2)
deldir 1.0-6 2021-10-23 [1] RSPM (R 4.2.0)
digest 0.6.31 2022-12-11 [1] RSPM (R 4.2.0)
dplyr * 1.1.2 2023-04-20 [1] RSPM (R 4.2.2)
ellipsis 0.3.2 2021-04-29 [1] RSPM (R 4.2.0)
entropy 1.3.1 2021-10-02 [1] RSPM (R 4.2.0)
evaluate 0.20 2023-01-17 [1] RSPM (R 4.2.0)
fansi 1.0.4 2023-01-22 [1] RSPM (R 4.2.0)
farver 2.1.1 2022-07-06 [1] RSPM (R 4.2.0)
fastmap 1.1.1 2023-02-24 [1] RSPM (R 4.2.0)
fitdistrplus 1.1-11 2023-04-25 [1] RSPM (R 4.2.2)
FNN 1.1.3.2 2023-03-20 [1] RSPM (R 4.2.0)
forcats * 1.0.0 2023-01-29 [1] RSPM (R 4.2.0)
foreach 1.5.2 2022-02-02 [1] RSPM (R 4.2.0)
future 1.32.0 2023-03-07 [1] RSPM (R 4.2.0)
future.apply 1.10.0 2022-11-05 [1] RSPM (R 4.2.0)
generics 0.1.3 2022-07-05 [1] RSPM (R 4.2.0)
GenomeInfoDb * 1.34.9 2023-02-02 [1] RSPM (R 4.2.2)
GenomeInfoDbData 1.2.9 2023-04-30 [1] RSPM (R 4.2.2)
GenomicRanges * 1.50.2 2022-12-16 [1] RSPM (R 4.2.2)
ggbeeswarm 0.7.2 2023-04-30 [1] Github (eclarke/ggbeeswarm@3cf58a9)
ggforce 0.4.1.9000 2023-04-30 [1] Github (thomasp85/ggforce@9be635c)
ggplot2 * 3.4.2 2023-04-03 [1] RSPM (R 4.2.0)
ggprism 1.0.4 2023-04-30 [1] Github (csdaw/ggprism@0e411f4)
ggrastr 1.0.1 2023-04-30 [1] Github (VPetukhov/ggrastr@7aed9af)
ggrepel 0.9.2.9999 2023-04-30 [1] Github (slowkow/ggrepel@fe3b5c3)
ggridges 0.5.4 2022-09-26 [1] RSPM (R 4.2.0)
ggsci 3.0.0 2023-04-30 [1] Github (nanxstats/ggsci@028b373)
glmnet * 4.1-7 2023-03-23 [1] RSPM (R 4.2.0)
GlobalOptions 0.1.2 2020-06-10 [1] RSPM (R 4.2.0)
globals 0.16.2 2022-11-21 [1] RSPM (R 4.2.0)
glue 1.6.2 2022-02-24 [1] RSPM (R 4.2.0)
goftest 1.2-3 2021-10-07 [1] RSPM (R 4.2.0)
gridExtra 2.3 2017-09-09 [1] RSPM (R 4.2.0)
gtable 0.3.3 2023-03-21 [1] RSPM (R 4.2.0)
hdf5r 1.3.8 2023-01-21 [1] RSPM (R 4.2.2)
here * 1.0.1 2020-12-13 [1] RSPM (R 4.2.0)
hexbin 1.28.3 2023-03-21 [1] RSPM (R 4.2.0)
hms 1.1.3 2023-03-21 [1] RSPM (R 4.2.0)
htmltools 0.5.5 2023-03-23 [1] RSPM (R 4.2.0)
htmlwidgets 1.6.2 2023-03-17 [1] RSPM (R 4.2.0)
httpuv 1.6.9 2023-02-14 [1] RSPM (R 4.2.0)
httr 1.4.5 2023-02-24 [1] RSPM (R 4.2.0)
ica 1.0-3 2022-07-08 [1] RSPM (R 4.2.0)
igraph 1.4.1 2023-02-24 [1] RSPM (R 4.2.0)
IRanges * 2.32.0 2022-11-01 [1] RSPM (R 4.2.2)
irlba 2.3.5.1 2022-10-03 [1] RSPM (R 4.2.0)
iterators 1.0.14 2022-02-05 [1] RSPM (R 4.2.0)
janitor 2.2.0.9000 2023-04-30 [1] Github (sfirke/janitor@d64c8bb)
jsonlite 1.8.4 2022-12-06 [1] RSPM (R 4.2.0)
KernSmooth 2.23-20 2021-05-03 [1] RSPM (R 4.2.0)
knitr 1.42 2023-01-25 [1] RSPM (R 4.2.0)
ks 1.14.0 2022-11-24 [1] RSPM (R 4.2.0)
labeling 0.4.2 2020-10-20 [1] RSPM (R 4.2.0)
later 1.3.0 2021-08-18 [1] RSPM (R 4.2.0)
lattice 0.21-8 2023-04-05 [1] RSPM (R 4.2.0)
lazyeval 0.2.2 2019-03-15 [1] RSPM (R 4.2.0)
leiden 0.4.3 2022-09-10 [1] RSPM (R 4.2.0)
lifecycle 1.0.3 2022-10-07 [1] RSPM (R 4.2.0)
liger 2.0.1 2023-04-30 [1] Github (JEFworks/liger@406dfa4)
listenv 0.9.0 2022-12-16 [1] RSPM (R 4.2.0)
lmtest 0.9-40 2022-03-21 [1] RSPM (R 4.2.0)
lubridate * 1.9.2 2023-02-10 [1] RSPM (R 4.2.0)
magrittr 2.0.3 2022-03-30 [1] RSPM (R 4.2.0)
MASS 7.3-58.1 2022-08-03 [1] CRAN (R 4.2.2)
Matrix * 1.5-4 2023-04-04 [1] RSPM (R 4.2.0)
MatrixGenerics * 1.10.0 2022-11-01 [1] RSPM (R 4.2.2)
matrixStats * 0.63.0 2022-11-18 [1] RSPM (R 4.2.0)
mclust 6.0.0 2022-10-31 [1] RSPM (R 4.2.0)
mgcv 1.8-42 2023-03-02 [1] RSPM (R 4.2.0)
mime 0.12 2021-09-28 [1] RSPM (R 4.2.0)
miniUI 0.1.1.1 2018-05-18 [1] RSPM (R 4.2.0)
munsell 0.5.0 2018-06-12 [1] RSPM (R 4.2.0)
mvtnorm 1.1-3 2021-10-08 [1] RSPM (R 4.2.0)
Nebulosa * 1.8.0 2022-11-01 [1] RSPM (R 4.2.2)
nlme 3.1-162 2023-01-31 [1] RSPM (R 4.2.0)
NNLM 0.4.4 2023-04-30 [1] Github (linxihui/NNLM@4574bca)
openssl 2.0.6 2023-03-09 [1] RSPM (R 4.2.0)
paletteer 1.5.0 2022-10-19 [1] RSPM (R 4.2.0)
parallelly 1.35.0 2023-03-23 [1] RSPM (R 4.2.0)
patchwork * 1.1.2.9000 2023-04-30 [1] Github (thomasp85/patchwork@c14c960)
pbapply 1.7-0 2023-01-13 [1] RSPM (R 4.2.0)
pillar 1.9.0 2023-03-22 [1] RSPM (R 4.2.0)
pkgconfig 2.0.3 2019-09-22 [1] RSPM (R 4.2.0)
plotly 4.10.1 2022-11-07 [1] RSPM (R 4.2.0)
plyr 1.8.8 2022-11-11 [1] RSPM (R 4.2.0)
png 0.1-8 2022-11-29 [1] RSPM (R 4.2.0)
polyclip 1.10-4 2022-10-20 [1] RSPM (R 4.2.0)
pracma 2.4.2 2022-09-22 [1] RSPM (R 4.2.0)
prismatic 1.1.1 2022-08-15 [1] RSPM (R 4.2.0)
progressr 0.13.0 2023-01-10 [1] RSPM (R 4.2.0)
promises 1.2.0.1 2021-02-11 [1] RSPM (R 4.2.0)
proxy 0.4-27 2022-06-09 [1] RSPM (R 4.2.0)
purrr * 1.0.1 2023-01-10 [1] RSPM (R 4.2.0)
R.methodsS3 1.8.2 2022-06-13 [1] RSPM (R 4.2.0)
R.oo 1.25.0 2022-06-12 [1] RSPM (R 4.2.0)
R.utils 2.12.2 2022-11-11 [1] RSPM (R 4.2.0)
R6 2.5.1 2021-08-19 [1] RSPM (R 4.2.0)
RANN 2.6.1 2019-01-08 [1] RSPM (R 4.2.0)
RColorBrewer * 1.1-3 2022-04-03 [1] RSPM (R 4.2.0)
Rcpp 1.0.10 2023-01-22 [1] RSPM (R 4.2.0)
RcppAnnoy 0.0.20 2022-10-27 [1] RSPM (R 4.2.0)
RCurl 1.98-1.12 2023-03-27 [1] RSPM (R 4.2.0)
readr * 2.1.4 2023-02-10 [1] RSPM (R 4.2.0)
rematch2 2.1.2 2020-05-01 [1] RSPM (R 4.2.0)
remotes 2.4.2 2021-11-30 [1] RSPM (R 4.2.0)
repr 1.1.6 2023-01-26 [1] RSPM (R 4.2.0)
reshape2 1.4.4 2020-04-09 [1] RSPM (R 4.2.0)
reticulate 1.28-9000 2023-04-30 [1] Github (rstudio/reticulate@442c49f)
rlang 1.1.0 2023-03-14 [1] RSPM (R 4.2.0)
rmarkdown 2.21 2023-03-26 [1] RSPM (R 4.2.0)
ROCR 1.0-11 2020-05-02 [1] RSPM (R 4.2.0)
rprojroot 2.0.3 2022-04-02 [1] RSPM (R 4.2.0)
RSpectra 0.16-1 2022-04-24 [1] RSPM (R 4.2.0)
rsvd 1.0.5 2021-04-16 [1] RSPM (R 4.2.0)
Rtsne 0.16 2022-04-17 [1] RSPM (R 4.2.0)
S4Vectors * 0.36.2 2023-02-26 [1] RSPM (R 4.2.2)
scales 1.2.1 2022-08-20 [1] RSPM (R 4.2.0)
scattermore 0.8 2022-02-14 [1] RSPM (R 4.2.0)
scCustomize * 1.1.1 2023-04-30 [1] Github (samuel-marsh/scCustomize@d08268d)
schex * 1.12.0 2022-11-01 [1] RSPM (R 4.2.2)
sctransform 0.3.5 2022-09-21 [1] RSPM (R 4.2.0)
sessioninfo 1.2.2 2021-12-06 [1] RSPM (R 4.2.0)
Seurat * 4.3.0 2022-11-18 [1] RSPM (R 4.2.2)
SeuratDisk * 0.0.0.9020 2023-04-30 [1] Github (mojaveazure/seurat-disk@9b89970)
SeuratObject * 4.1.3 2022-11-07 [1] RSPM (R 4.2.0)
SeuratWrappers * 0.3.1 2023-04-30 [1] Github (satijalab/seurat-wrappers@d28512f)
shape 1.4.6 2021-05-19 [1] RSPM (R 4.2.0)
shiny * 1.7.4 2022-12-15 [1] RSPM (R 4.2.0)
SingleCellExperiment * 1.20.1 2023-03-17 [1] RSPM (R 4.2.2)
skimr 2.1.5 2023-04-30 [1] Github (ropensci/skimr@d5126aa)
snakecase 0.11.0 2019-05-25 [1] RSPM (R 4.2.0)
snow 0.4-4 2021-10-27 [1] RSPM (R 4.2.0)
sp 1.6-0 2023-01-19 [1] RSPM (R 4.2.0)
spatstat.data 3.0-1 2023-03-12 [1] RSPM (R 4.2.0)
spatstat.explore 3.1-0 2023-03-14 [1] RSPM (R 4.2.0)
spatstat.geom 3.1-0 2023-03-12 [1] RSPM (R 4.2.0)
spatstat.random 3.1-4 2023-03-13 [1] RSPM (R 4.2.0)
spatstat.sparse 3.0-1 2023-03-12 [1] RSPM (R 4.2.0)
spatstat.utils 3.0-2 2023-03-11 [1] RSPM (R 4.2.0)
stringi 1.7.12 2023-01-11 [1] RSPM (R 4.2.0)
stringr * 1.5.0 2022-12-02 [1] RSPM (R 4.2.0)
SummarizedExperiment * 1.28.0 2022-11-01 [1] RSPM (R 4.2.2)
survival 3.5-5 2023-03-12 [1] RSPM (R 4.2.0)
swne * 0.6.20 2023-04-30 [1] Github (yanwu2014/swne@05fc3ee)
systemfonts 1.0.4 2022-02-11 [1] RSPM (R 4.2.0)
tensor 1.5 2012-05-05 [1] RSPM (R 4.2.0)
tibble * 3.2.1 2023-03-20 [1] RSPM (R 4.2.0)
tidyr * 1.3.0 2023-01-24 [1] RSPM (R 4.2.0)
tidyselect 1.2.0 2022-10-10 [1] RSPM (R 4.2.0)
tidyverse * 2.0.0.9000 2023-04-30 [1] Github (tidyverse/tidyverse@8ec2e1f)
timechange 0.2.0 2023-01-11 [1] RSPM (R 4.2.0)
tweenr 2.0.2 2022-09-06 [1] RSPM (R 4.2.0)
tzdb 0.3.0 2022-03-28 [1] RSPM (R 4.2.0)
umap 0.2.10.0 2023-02-01 [1] RSPM (R 4.2.0)
UpSetR * 1.4.0 2023-04-30 [1] Github (hms-dbmi/UpSetR@b14854a)
usedist 0.4.0 2020-03-01 [1] RSPM (R 4.2.0)
utf8 1.2.3 2023-01-31 [1] RSPM (R 4.2.0)
uwot 0.1.14 2022-08-22 [1] RSPM (R 4.2.0)
V8 4.2.2 2022-11-03 [1] RSPM (R 4.2.0)
vctrs 0.6.2 2023-04-19 [1] RSPM (R 4.2.2)
vipor 0.4.5 2017-03-22 [1] RSPM (R 4.2.0)
viridis 0.6.2 2021-10-13 [1] RSPM (R 4.2.0)
viridisLite 0.4.1 2022-08-22 [1] RSPM (R 4.2.0)
withr 2.5.0 2022-03-03 [1] RSPM (R 4.2.0)
xfun 0.39 2023-04-20 [1] RSPM (R 4.2.2)
xtable 1.8-4 2019-04-21 [1] RSPM (R 4.2.0)
XVector 0.38.0 2022-11-01 [1] RSPM (R 4.2.2)
yaml 2.3.7 2023-01-23 [1] RSPM (R 4.2.0)
zlibbioc 1.44.0 2022-11-01 [1] RSPM (R 4.2.2)
zoo 1.8-12 2023-04-13 [1] RSPM (R 4.2.2)
[1] /opt/R/4.2.2/lib/R/library
──────────────────────────────────────────────────────────────────────────────